I am having a problem with subsetting. When I subset my data set, several columns in the resulting subset are filled with 0's and the variable class for these columns has changed to unknown. This happens consistently with certain subsets. The columns affected vary between affected subsets
I don't understand why this is happening. All I am doing is a simple subset command. Why is R losing 4 whole columns of numerical data and replacing it with nonsense.
The offending piece of code is this simple command here:
table.al = subset(bamboo_compounds,bamboo_compounds$CClass=="aldehyde")
The original data set looks like this:
The resulting subset looks like this:
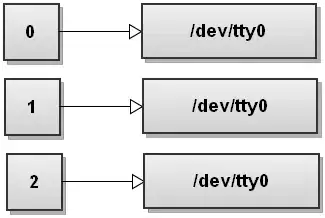
Those four columns should be filled with numerical data.
I have literally done nothing other than load in a .csv file and then make a subset of that data. Please, can someone give me some idea of might be causing this and how I can avoid it?