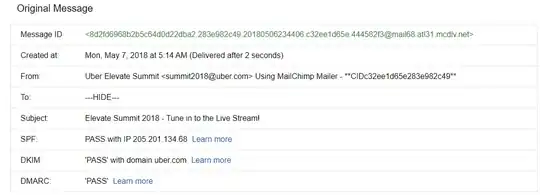
I have a folder containing several thousand .txt files. I'd like to combine them in a big .csv according to the following model:
I found a R script supposed to do the job (https://gist.github.com/benmarwick/9265414), but it displays this error.
Error in read.table(file = file, header = header, sep = sep, quote = quote, : duplicate 'row.names' are not allowed
I don't understand what's my mistake.
No matter, I'm pretty sure there's a way to do that without R. If you know a very elegant and simple one, it would be appreciated (and useful for a lot of guys like me)
PRECISION : the text files are in french, so not ASCII. Here is a sample : https://www.dropbox.com/s/rj4df94hqisod5z/Texts.zip?dl=0