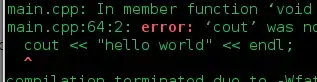
I would like to plot both a linear model (LM) and non-linear (GLM) model of the same data.
The range between 16% - 84% should line up between a LM and GLM, Citation: section 3.5
I have included a more complete chunk of the code because I am not sure at which point I should try to cut the linear model. or at which point I have messed up - I think with the linear model.
The code below results in the following image:
My Objective (taken from previous citation-link).
Here is my data:
mydata3 <- structure(list(
dose = c(0, 0, 0, 3, 3, 3, 7.5, 7.5, 7.5, 10, 10, 10, 25, 25, 25, 50, 50, 50),
total = c(25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L, 25L),
affected = c(1, 0, 1.2, 2.8, 4.8, 9, 2.8, 12.8, 8.6, 4.8, 4.4, 10.2, 6, 20, 14, 12.8, 23.4, 21.6),
probability = c(0.04, 0, 0.048, 0.112, 0.192, 0.36, 0.112, 0.512, 0.344, 0.192, 0.176, 0.408, 0.24, 0.8, 0.56, 0.512, 0.936, 0.864)),
.Names = c("dose", "total", "affected", "probability"),
row.names = c(NA, -18L),
class = "data.frame")
My script:
#load libraries
library(ggplot2)
library(drc) # glm model
library(plyr) # rename function
library(scales) #log plot scale
#Creating linear model
mod_linear <- lm(probability ~ (dose), weights = total, data = mydata3)
#Creating data.frame: note values 3 and 120 refer to 16% and 84% response in sigmoidal plot
line_df <-expand.grid(dose=exp(seq(log(3),log(120),length=200)))
#Extracting values from linear model
p_line_df <- as.data.frame(cbind(dose = line_df,
predict(mod_linear, newdata=data.frame(dose = line_df),
interval="confidence",level=0.95)))
#Renaming linear df columns
p_line_df <-rename(p_line_df, c("fit"="probability"))
p_line_df <-rename(p_line_df, c("lwr"="Lower"))
p_line_df <-rename(p_line_df, c("upr"="Upper"))
p_line_df$model <-"Linear"
#Create sigmoidal dose-response curve using drc package
mod3 <- drm(probability ~ (dose), weights = total, data = mydata3, type ="binomial", fct=LL.2(names=c("Slope:b","ED50:e")))
#data frame for ggplot2
base_DF_3 <-expand.grid(dose=exp(seq(log(1.0000001),log(10000),length=200)))
#extract data from model
p_df3 <- as.data.frame(cbind(dose = base_DF_3,
predict(mod3, newdata=data.frame(dose = base_DF_3),
interval="confidence", level=.95)))
#renaming columns
p_df3 <-rename(p_df3, c("Prediction"="probability"))
p_df3$model <-"Sigmoidal"
#combining Both DataFames
p_df_all <- rbind(p_df3, p_line_df)
#plotting
ggplot(p_df_all, aes(x=dose,y=probability, group=model))+
geom_line(aes(x=dose,y=probability,group=model,linetype=model),show.legend = TRUE)+
scale_x_log10(breaks = c(0.000001, 10^(0:10)),labels = c(0, math_format()(0:10)))