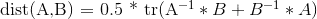Here is a humble optimisation, saving 9 multiplications and 3 subtractions
def inversion(m):
m1, m2, m3, m4, m5, m6, m7, m8, m9 = m.ravel()
inv = np.array([[m5*m9-m6*m8, m3*m8-m2*m9, m2*m6-m3*m5],
[m6*m7-m4*m9, m1*m9-m3*m7, m3*m4-m1*m6],
[m4*m8-m5*m7, m2*m7-m1*m8, m1*m5-m2*m4]])
return inv / np.dot(inv[0], m[:, 0])
You can squeeze out a few more ops (another 24 multiplications if I'm counting correctly) by doing the entire trace in one go:
def det(m):
m1, m2, m3, m4, m5, m6, m7, m8, m9 = m.ravel()
return np.dot(m[:, 0], [m5*m9-m6*m8, m3*m8-m2*m9, m2*m6-m3*m5])
# or try m1*(m5*m9-m6*m8) + m4*(m3*m8-m2*m9) + m7*(m2*m6-m3*m5)
# probably the fastest would be to inline the two calls to det
# I'm not doing it here because of readability but you should try it
def dist(m, n):
m1, m2, m3, m4, m5, m6, m7, m8, m9 = m.ravel()
n1, n2, n3, n4, n5, n6, n7, n8, n9 = n.ravel()
return 0.5 * np.dot(
m.ravel()/det(m) + n.ravel()/det(n),
[m5*n9-m6*n8, m6*n7-m4*n9, m4*n8-m5*n7, n3*m8-n2*m9, n1*m9-n3*m7,
n2*m7-n1*m8, m2*n6-m3*n5, m3*n4-m1*n6, m1*n5-m2*n4])
Ok here is the inlined version:
import numpy as np
from timeit import timeit
def dist(m, n):
m1, m2, m3, m4, m5, m6, m7, m8, m9 = m.ravel()
n1, n2, n3, n4, n5, n6, n7, n8, n9 = n.ravel()
return 0.5 * np.dot(
m.ravel()/(m1*(m5*m9-m6*m8) + m4*(m3*m8-m2*m9) + m7*(m2*m6-m3*m5))
+ n.ravel()/(n1*(n5*n9-n6*n8) + n4*(n3*n8-n2*n9) + n7*(n2*n6-n3*n5)),
[m5*n9-m6*n8, m6*n7-m4*n9, m4*n8-m5*n7, n3*m8-n2*m9, n1*m9-n3*m7,
n2*m7-n1*m8, m2*n6-m3*n5, m3*n4-m1*n6, m1*n5-m2*n4])
def dist_np(m, n):
return 0.5 * np.diag(np.linalg.inv(m)@n + np.linalg.inv(n)@m).sum()
for i in range(3):
A, B = np.random.random((2,3,3))
print(dist(A, B), dist_np(A, B))
print('pp ', timeit('f(A,B)', number=10000, globals={'f':dist, 'A':A, 'B':B}))
print('numpy ', timeit('f(A,B)', number=10000, globals={'f':dist_np, 'A':A, 'B':B}))
prints:
2.20109953156 2.20109953156
pp 0.13215381593909115
numpy 0.4334693900309503
7.50799877993 7.50799877993
pp 0.13934064202476293
numpy 0.32861811900511384
-0.780284449609 -0.780284449609
pp 0.1258618349675089
numpy 0.3110764700686559
Note that you can make another substantial saving by batch-processing using a vectorised version of the function. The test computes all 10,000 pairwise distances between two batches of 100 matrices:
def dist(m, n):
m = np.moveaxis(np.reshape(m, m.shape[:-2] + (-1,)), -1, 0)
n = np.moveaxis(np.reshape(n, n.shape[:-2] + (-1,)), -1, 0)
m1, m2, m3, m4, m5, m6, m7, m8, m9 = m
n1, n2, n3, n4, n5, n6, n7, n8, n9 = n
return 0.5 * np.einsum("i...,i...->...",
m/(m1*(m5*m9-m6*m8) + m4*(m3*m8-m2*m9) + m7*(m2*m6-m3*m5))
+ n/(n1*(n5*n9-n6*n8) + n4*(n3*n8-n2*n9) + n7*(n2*n6-n3*n5)),
[m5*n9-m6*n8, m6*n7-m4*n9, m4*n8-m5*n7, n3*m8-n2*m9, n1*m9-n3*m7,
n2*m7-n1*m8, m2*n6-m3*n5, m3*n4-m1*n6, m1*n5-m2*n4])
def dist_np(m, n):
return 0.5 * (np.linalg.inv(m)@n + np.linalg.inv(n)@m)[..., np.arange(3), np.arange(3)].sum(axis=-1)
for i in range(3):
A = np.random.random((100,1,3,3))
B = np.random.random((1,100,3,3))
print(np.allclose(dist(A, B), dist_np(A, B)))
print('pp ', timeit('f(A,B)', number=100, globals={'f':dist, 'A':A, 'B':B}))
print('numpy ', timeit('f(A,B)', number=100, globals={'f':dist_np, 'A':A, 'B':B}))
prints:
True
pp 0.14652886800467968
numpy 1.5294789629988372
True
pp 0.1482033939100802
numpy 1.6455406049499288
True
pp 0.1279512889450416
numpy 1.370200254023075