aweek::get_date allows you to get weekly dates only using the year and epiweek.
Here I created a reprex with a sequence of dates (link), extract the epiweek with lubridate::epiweek, defined sunday as start of a week with aweek::set_week_start, summarized weekly values, created a new date vector with aweek::get_date, and plot them.
library(tidyverse)
library(lubridate)
library(aweek)
data_ts <- tibble(date=seq(ymd('2012-04-07'),
ymd('2014-03-22'),
by = '1 day')) %>%
mutate(value = rnorm(n(),mean = 5),
#using aweek
epidate=date2week(date,week_start = 7),
#using lubridate
epiweek=epiweek(date),
dayw=wday(date,label = T,abbr = F),
month=month(date,label = F,abbr = F),
year=year(date)) %>%
print()
#> # A tibble: 715 x 7
#> date value epidate epiweek dayw month year
#> <date> <dbl> <aweek> <dbl> <ord> <dbl> <dbl>
#> 1 2012-04-07 3.54 2012-W14-7 14 sábado 4 2012
#> 2 2012-04-08 5.79 2012-W15-1 15 domingo 4 2012
#> 3 2012-04-09 4.50 2012-W15-2 15 lunes 4 2012
#> 4 2012-04-10 5.44 2012-W15-3 15 martes 4 2012
#> 5 2012-04-11 5.13 2012-W15-4 15 miércoles 4 2012
#> 6 2012-04-12 4.87 2012-W15-5 15 jueves 4 2012
#> 7 2012-04-13 3.28 2012-W15-6 15 viernes 4 2012
#> 8 2012-04-14 5.72 2012-W15-7 15 sábado 4 2012
#> 9 2012-04-15 6.91 2012-W16-1 16 domingo 4 2012
#> 10 2012-04-16 4.58 2012-W16-2 16 lunes 4 2012
#> # ... with 705 more rows
#CORE: Here you set the start of the week!
set_week_start(7) #sunday
get_week_start()
#> [1] 7
data_ts_w <- data_ts %>%
group_by(year,epiweek) %>%
summarise(sum_week_value=sum(value)) %>%
ungroup() %>%
#using aweek
mutate(epi_date=get_date(week = epiweek,year = year),
wik_date=date2week(epi_date)
) %>%
print()
#> # A tibble: 104 x 5
#> year epiweek sum_week_value epi_date wik_date
#> <dbl> <dbl> <dbl> <date> <aweek>
#> 1 2012 1 11.0 2012-01-01 2012-W01-1
#> 2 2012 14 3.54 2012-04-01 2012-W14-1
#> 3 2012 15 34.7 2012-04-08 2012-W15-1
#> 4 2012 16 35.1 2012-04-15 2012-W16-1
#> 5 2012 17 34.5 2012-04-22 2012-W17-1
#> 6 2012 18 34.7 2012-04-29 2012-W18-1
#> 7 2012 19 36.5 2012-05-06 2012-W19-1
#> 8 2012 20 32.1 2012-05-13 2012-W20-1
#> 9 2012 21 35.4 2012-05-20 2012-W21-1
#> 10 2012 22 37.5 2012-05-27 2012-W22-1
#> # ... with 94 more rows
#you can use get_date output with ggplot
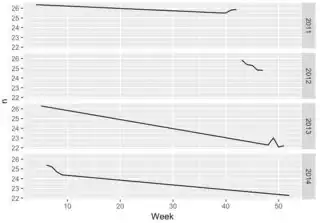
data_ts_w %>%
slice(-(1:3)) %>%
ggplot(aes(epi_date, sum_week_value)) +
geom_line() +
scale_x_date(date_breaks="5 week", date_labels = "%Y-%U") +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
labs(title = "Weekly time serie",
x="Time (Year - CDC epidemiological week)",
y="Sum of weekly values")
ggsave("figure/000-timeserie-week.png",height = 3,width = 10)
Created on 2019-08-12 by the reprex package (v0.3.0)