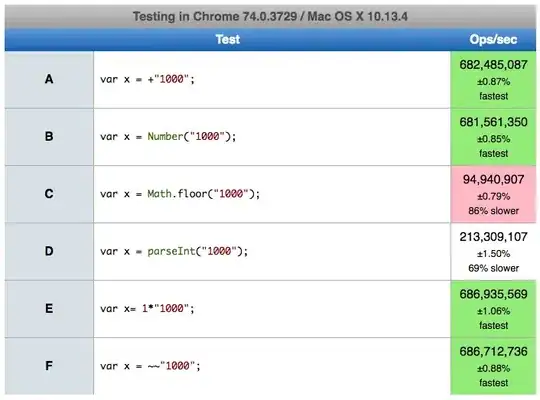
For this type of plot I would construct a data set to be plotted by geom_rect. For an example, the data set below is constructed such that the ymin and ymax of a rectangle are defined by the order of the ACGT at each position. The graphic below may not look much like the one that you have in your question, but the method provided should produce a similar graphic given your data set. You may need to tweak geom_text values and colors, but I think the primary question to answer is how to construct a data set and plot it.
library(ggplot2)
library(dplyr)
library(tidyr)
# Make some random data
set.seed(42)
rna_seq_data <-
data_frame(position = 1:25,
A = floor(runif(25, 0, 5000)),
C = floor(runif(25, 0, 5000)),
G = floor(runif(25, 0, 5000)),
T = floor(runif(25, 0, 5000)))
tidyr::gather(rna_seq_data, key, value, -position) %>%
dplyr::group_by(position) %>%
dplyr::mutate(order = rank(value)) %>%
dplyr::arrange(position, order) %>%
dplyr::mutate(ymin = dplyr::if_else(order == 1, 0, lag(value)),
ymax = cumsum(value),
xmin = position - 0.45,
xmax = position + 0.45) %>%
dplyr::ungroup() %>%
ggplot(.) +
aes(xmin = xmin, xmax = xmax, ymin = ymin, ymax = ymax, fill = key) +
geom_rect()