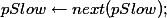I am a fan of data.table, as of writing re-usable functions for all current and future needs.
Here's a challenge I run into while working on the answer to this problem: Best way to plot automatically all data.table columns using ggplot2
We pass data.table to a function for plotting and then the original data.table gets modified, even though we made a copy of it to prevent that.
Here's a simple code to illustrate:
plotYofX <- function(.dt,x,y) {
dt <- .dt
dt[, (c(x,y)) := lapply(.SD, function(x) {as.numeric(x)}), .SDcols = c(x,y)]
ggplot(dt) + geom_step(aes(x=get(names(dt)[x]), y=get(names(dt)[y]))) + labs(x=names(dt)[x], y=names(dt)[y])
}
> dtDiamonds <- data.table(ggplot2::diamonds[2:5,1:3]);
> dtDiamonds
carat cut color
<num> <ord> <ord>
1: 0.21 Premium E
2: 0.23 Good E
3: 0.29 Premium I
4: 0.31 Good J
> plotYofX(dtDiamonds,1,2);
> dtDiamonds
carat cut color
<num> <num> <ord>
1: 0.21 4 E
2: 0.23 2 E
3: 0.29 4 I
4: 0.31 2 J
I've seen many postings on various issues related to using := inside the function, but could not find any to help me to resolve this seemingly very easy issue. (Of course, I don't what to convert it back to data.frame to achieve the desired outcome)