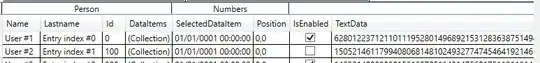
I would start like this:
- binarize image
- find each set pixel of tube
flood fill that position by tube color
use any filling algorithm with 8-neighbors and during filling also count recolored pixels in some counter cnt.
if area size cnt too small recolor it to background otherwise account its size cnt/average_tube_width to the histogram.
Here simple C++ example of this:
picture pic0,pic1;
// pic0 - source img
// pic1 - output img
// 0xAARRGGBB
const DWORD col_backg=0x00202020; // gray
const DWORD col_tube =0x00FFFFFF; // white
const DWORD col_done =0x0000A0FF; // aqua
const DWORD col_noise=0x00000080; // blue
const DWORD col_error=0x00FF0000; // red (too smal _hist value)
const DWORD col_hist =0x00FFFF00; // yellow
const DWORD col_test =0x01000000; // A* filling start color (must be bigger then all other colors used)
int x,y,xx,yy,i;
DWORD c;
const int _hist=256; // max area size for histogram
int hist[_hist]; // histogram
// copy source image to output
pic1=pic0;
pic1.enhance_range(); // maximize dynamic range <0,255>^3
pic1.pixel_format(_pf_u); // convert to grayscale <0,765>
pic1.threshold(100,766,col_backg,col_tube); // threshold intensity to binarize image
pic1.pf=_pf_rgba; // set as RGBA (without conversion)
// clear histogram
for (i=0;i<_hist;i++) hist[i]=0;
// find all tubes
for (y=0;y<pic1.ys;y++)
for (x=0;x<pic1.xs;x++)
if (pic1.p[y][x].dd==col_tube)
{
pic1.Astarfill(x,y,col_test); // fill count area (8 neighbors)
if (pic1._floodfill_n>5) // if valid size
{
c=col_done; // set recolor color to done
// update histogram
if (pic1._floodfill_n<_hist) hist[pic1._floodfill_n]++;
else c=col_error;
}
else c=col_noise;
// recolor filled bbox with c
for (yy=pic1._floodfill_y0;yy<=pic1._floodfill_y1;yy++)
for (xx=pic1._floodfill_x0;xx<=pic1._floodfill_x1;xx++)
if (pic1.p[yy][xx].dd>=col_test)
pic1.p[yy][xx].dd=c;
}
// render histogram
for (x=0;x<_hist;x++)
for (i=0,y=pic1.ys-1;(y>=0)&&(i<hist[x]<<2);y--,i++)
pic1.p[y][x].dd=col_hist;
The result for your input image is:
The yellow lines are the lengths distribution (x axis is tube length and y is probability)
I use my own picture class for images so some members are:
xs,ys is size of image in pixels
p[y][x].dd is pixel at (x,y) position as 32 bit integer type
clear(color) clears entire image with color
resize(xs,ys) resizes image to new resolution
bmp is VCL encapsulated GDI Bitmap with Canvas access
pf holds actual pixel format of the image:
enum _pixel_format_enum
{
_pf_none=0, // undefined
_pf_rgba, // 32 bit RGBA
_pf_s, // 32 bit signed int
_pf_u, // 32 bit unsigned int
_pf_ss, // 2x16 bit signed int
_pf_uu, // 2x16 bit unsigned int
_pixel_format_enum_end
};
color and pixels are encoded like this:
union color
{
DWORD dd; WORD dw[2]; byte db[4];
int i; short int ii[2];
color(){}; color(color& a){ *this=a; }; ~color(){}; color* operator = (const color *a) { dd=a->dd; return this; }; /*color* operator = (const color &a) { ...copy... return this; };*/
};
The bands are:
enum{
_x=0, // dw
_y=1,
_b=0, // db
_g=1,
_r=2,
_a=3,
_v=0, // db
_s=1,
_h=2,
};
I also use mine dynamic list template so:
List<double> xxx; is the same as double xxx[];
xxx.add(5); adds 5 to end of the list
xxx[7] access array element (safe)
xxx.dat[7] access array element (unsafe but fast direct access)
xxx.num is the actual used size of the array
xxx.reset() clears the array and set xxx.num=0
xxx.allocate(100) preallocate space for 100 items
Now the A* filling is implemented like this:
// these are picture:: members to speed up recursive fillings
int _floodfill_rn; // anti stack overflow recursions
List<int> _floodfill_xy; // anti stack overflow pendng recursions
int _floodfill_a0[4]; // recursion filled color and fill color
color _floodfill_c0,_floodfill_c1; // recursion filled color and fill color
int _floodfill_x0,_floodfill_x1,_floodfill_n; // recursion bounding box and filled pixel count
int _floodfill_y0,_floodfill_y1;
// here the filling I used
void picture::Astarfill(int x,int y,DWORD id)
{
_floodfill_c0=p[y][x];
_floodfill_c1.dd=id;
_floodfill_n=0;
_floodfill_x0=x;
_floodfill_y0=y;
_floodfill_x1=x;
_floodfill_y1=y;
_floodfill_rn=0;
_floodfill_xy.num=0;
if ((x<0)||(x>=xs)||(y<0)||(y>=ys)) return;
int i;
List<int> xy0,xy1,*p0,*p1,*pp;
// first point
p0=&xy0;
p1=&xy1;
p0->num=0;
p0->add(x); p0->add(y); p[y][x].dd=id; _floodfill_n++;
for (;p0->num;)
{
p1->num=0; id++;
for (i=0;i<p0->num;)
{
x=p0->dat[i]; i++;
y=p0->dat[i]; i++;
x--; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
y--; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
x++; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
x++; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
y++; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
y++; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
x--; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
x--; if ((x>=0)&&(y>=0)&&(x<xs)&&(y<ys)&&(p[y][x].dd==_floodfill_c0.dd)){ p1->add(x); p1->add(y); p[y][x].dd=id; _floodfill_n++; if (_floodfill_x0>x) _floodfill_x0=x; if (_floodfill_y0>y) _floodfill_y0=y; if (_floodfill_x1<x) _floodfill_x1=x; if (_floodfill_y1<y) _floodfill_y1=y; }
}
pp=p0; p0=p1; p1=pp;
}
_floodfill_rn=id-1;
}
If you want to improve your counting based on the size then if you got a multiple of avg size you got intersected tubes. So you can either try to compute how many of them are there and account the avg size to the histogram instead of using the full size or us A* filling and locate the endpoints. If you find more than 2 endpoints you can try to distinct between tubes.
So first use A* fill to find local max and then A* fill again from that position (so you start filling from endpoint). Then find all local maxs and based on avg size and actual size of tube and number of endpoints you can determine how many tubes are grouped together and how many of them are interconnected. Then you can try to do all possible combinations between endpoints and the one closest to avg size of tube per each tube is the most "correct" one. That should enhance precision a lot more.
In case you do not know the avg tube thickness you can use A* filling for non intersecting tubes directly to acquire the length. So in the second fill (from endpoint) when the filling stops the last filled ID is the length of tube in pixels.