I have a dataframe like so (the real one has 300+ rows):
cline endpt fx type colours
SF-268 96.5 1 CNS #848B9E
22 SF-268 103.3 2 CNS #848B9E
23 SF-268 60.7 3 CNS #848B9E
24 SF-268 5.0 4 CNS #848B9E
25 SF-268 8.7 5 CNS #848B9E
26 SF-268 -9.4 6 CNS #848B9E
27 SF-268 -20.7 7 CNS #848B9E
28 SNB-75 105.5 1 CNS #848B9E
29 SNB-75 94.5 2 CNS #848B9E
30 SNB-75 35.3 3 CNS #848B9E
.. ... ... .. ... ...
71 SW-620 95.6 2 Colon #468F14
72 SW-620 73.5 3 Colon #468F14
73 SW-620 4.0 4 Colon #468F14
74 SW-620 9.7 5 Colon #468F14
75 SW-620 -58.6 6 Colon #468F14
76 SW-620 -49.1 7 Colon #468F14
77 CCRF-CEM 95.8 1 Leukemia #FF041E
78 CCRF-CEM 96.6 2 Leukemia #FF041E
79 CCRF-CEM 89.2 3 Leukemia #FF041E
80 CCRF-CEM 3.5 4 Leukemia #FF041E
81 CCRF-CEM 13.7 5 Leukemia #FF041E
82 CCRF-CEM -21.3 6 Leukemia #FF041E
83 CCRF-CEM -6.6 7 Leukemia #FF041E
84 HL-60(TB) 93.9 1 Leukemia #FF041E
85 HL-60(TB) 95.3 2 Leukemia #FF041E
86 HL-60(TB) 94.0 3 Leukemia #FF041E
87 HL-60(TB) 13.3 4 Leukemia #FF041E
88 HL-60(TB) 14.6 5 Leukemia #FF041E
89 HL-60(TB) -44.0 6 Leukemia #FF041E
90 HL-60(TB) -57.0 7 Leukemia #FF041E
91 K-562 88.1 1 Leukemia #FF041E
92 K-562 97.1 2 Leukemia #FF041E
93 K-562 73.6 3 Leukemia #FF041E
94 K-562 6.6 4 Leukemia #FF041E
95 K-562 7.0 5 Leukemia #FF041E
96 K-562 -21.9 6 Leukemia #FF041E
97 K-562 -29.6 7 Leukemia #FF041E
98 MOLT-4 98.9 1 Leukemia #FF041E
99 MOLT-4 96.8 2 Leukemia #FF041E
100 MOLT-4 68.9 3 Leukemia #FF041E
I used the following examples to help me produce my code at the bottom:
- How to get different colored lines for different plots in a single figure?
- Plotting multiple lines with pandas dataframe
- plot different color for different categorical levels using matplotlib.
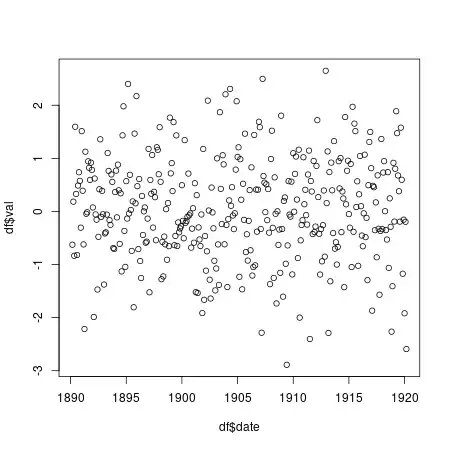
I managed to get a plot, however I think the line plot connects the last y value with the first, making a straight line (image below). I'm not sure why. Any help would be appreciated. Thanks.
import csv
import numpy as np
import pandas as pd
import itertools
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
labels = []
for key, grp in dfm.groupby(['colours']):
ax = grp.plot(ax=ax,linestyle='-',marker='s',x='fx',y='endpt',c=key)
labels.append(key)
lines, _ = ax.get_legend_handles_labels()
g=[]
for i in labels:
g.append(list(co.keys())[list(co.values()).index(i)])
ax.legend(lines, g, loc='best')