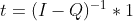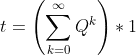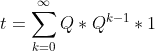I have a very large absorbing Markov chain. I want to obtain the fundamental matrix of this chain to calculate the expected number of steps before absortion. From this question I know that this can be calculated by the equation
(I - Q)t=1
which can be obtained by using the following python code:
def expected_steps_fast(Q):
I = numpy.identity(Q.shape[0])
o = numpy.ones(Q.shape[0])
numpy.linalg.solve(I-Q, o)
However, I would like to calculate it using some kind of iterative method similar to the power iteration method used for calculate the PageRank. This method would allow me to calculate an approximation to the expected number of steps before absortion in a mapreduce-like system.
¿Does something similar exist?