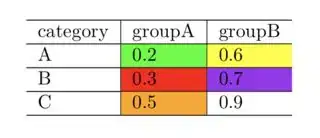
I cannot get cellcolor to work in rmarkdown:
---
header-includes:
- \usepackage{colortbl}
- \usepackage{color}
output:
pdf_document
---
```{r, results="asis"}
library(xtable)
# Your data
tab = data.frame(category = c("A","B","C"), groupA = c(.2,.3,.5), groupB= c(.6,.7,.9))
# Function to cut your data, and assign colour to each range
f <- function(x) cut(x, c(0, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, Inf),
labels=c("green", "red", "blue", "orange", "yellow", "purple", "brown", "white"),
include.lowest = FALSE, right = TRUE)
# Apply function to columns: this overwrites your data
tab[c("groupA", "groupB")] <- lapply(tab[c("groupA", "groupB")], function(x)
paste0("\\cellcolor{", f(x), "}", x))
# Sanitise output
print(xtable(tab), sanitize.text.function = identity)
```
I keep getting this error:
! Undefined control sequence.
l.155 1 & A & \cellcolor
Any ideas what does cellcolor needs to work?