Output examples of KFold, StratifiedKFold, StratifiedShuffleSplit:
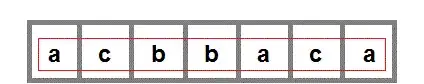
The above pictorial output is an extension of @Ken Syme's code:
from sklearn.model_selection import KFold, StratifiedKFold, StratifiedShuffleSplit
SEED = 43
SPLIT = 3
X_train = [0,1,2,3,4,5,6,7,8]
y_train = [0,0,0,0,0,0,1,1,1] # note 6,7,8 are labelled class '1'
print("KFold, shuffle=False (default)")
kf = KFold(n_splits=SPLIT, random_state=SEED)
for train_index, test_index in kf.split(X_train, y_train):
print("TRAIN:", train_index, "TEST:", test_index)
print("KFold, shuffle=True")
kf = KFold(n_splits=SPLIT, shuffle=True, random_state=SEED)
for train_index, test_index in kf.split(X_train, y_train):
print("TRAIN:", train_index, "TEST:", test_index)
print("\nStratifiedKFold, shuffle=False (default)")
skf = StratifiedKFold(n_splits=SPLIT, random_state=SEED)
for train_index, test_index in skf.split(X_train, y_train):
print("TRAIN:", train_index, "TEST:", test_index)
print("StratifiedKFold, shuffle=True")
skf = StratifiedKFold(n_splits=SPLIT, shuffle=True, random_state=SEED)
for train_index, test_index in skf.split(X_train, y_train):
print("TRAIN:", train_index, "TEST:", test_index)
print("\nStratifiedShuffleSplit")
sss = StratifiedShuffleSplit(n_splits=SPLIT, random_state=SEED, test_size=3)
for train_index, test_index in sss.split(X_train, y_train):
print("TRAIN:", train_index, "TEST:", test_index)
print("\nStratifiedShuffleSplit (can customise test_size)")
sss = StratifiedShuffleSplit(n_splits=SPLIT, random_state=SEED, test_size=2)
for train_index, test_index in sss.split(X_train, y_train):
print("TRAIN:", train_index, "TEST:", test_index)