I've packaged a solution in a one function github package.
I'm copying the whole code at the bottom but the simplest is :
remotes::install_github("moodymudskipper/datasearch")
library(datasearch)
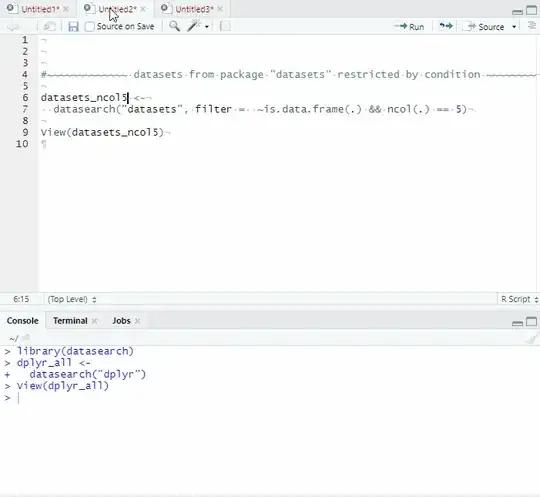
All data sets from package "dplyr"
dplyr_all <-
datasearch("dplyr")
View(dplyr_all)

Datasets from package "datasets" restricted by condition
datasets_ncol5 <-
datasearch("datasets", filter = ~is.data.frame(.) && ncol(.) == 5)
View(datasets_ncol5)
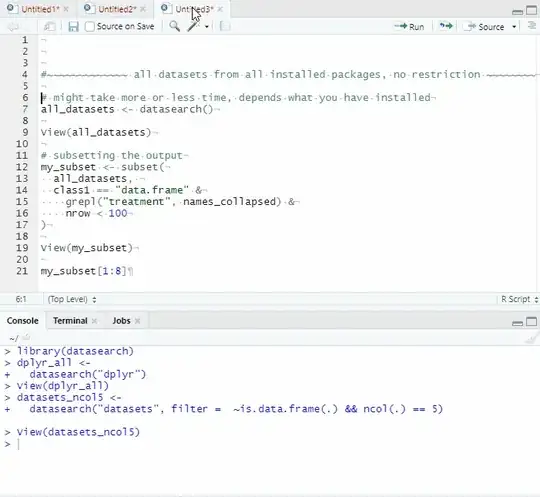
All datasets from all installed packages, no restriction
# might take more or less time, depends what you have installed
all_datasets <- datasearch()
View(all_datasets)
# subsetting the output
my_subset <- subset(
all_datasets,
class1 == "data.frame" &
grepl("treatment", names_collapsed) &
nrow < 100
)
View(my_subset)
datasearch <- function(pkgs = NULL, filter = NULL){
# make function silent
w <- options()$warn
options(warn = -1)
search_ <- search()
file_ <- tempfile()
file_ <- file(file_, "w")
on.exit({
options(warn = w)
to_detach <- setdiff(search(), search_)
for(pkg in to_detach) eval(bquote(detach(.(pkg))))
# note : we still have loaded namespaces, we could unload those that we ddn't
# have in the beginning but i'm worried about surprising effects, I think
# the S3 method tables should be cleaned too, and maybe other things
# note2 : tracing library and require didn't work
})
# convert formula to function
if(inherits(filter, "formula")) {
filter <- as.function(c(alist(.=), filter[[length(filter)]]))
}
## by default fetch all available packages in .libPaths()
if(is.null(pkgs)) pkgs <- .packages(all.available = TRUE)
## fetch all data sets description
df <- as.data.frame(data(package = pkgs, verbose = FALSE)$results)
names(df) <- tolower(names(df))
item <- NULL # for cmd check note
df <- transform(
df,
data_name = sub('.*\\((.*)\\)', '\\1', item),
dataset = sub(' \\(.*', '', item),
libpath = NULL,
item = NULL
)
df <- df[order(df$package, df$data_name),]
pkg_data_names <- aggregate(dataset ~ package + data_name, df, c)
pkg_data_names <- pkg_data_names[order(pkg_data_names$package, pkg_data_names$data_name),]
env <- new.env()
n <- nrow(pkg_data_names)
pb <- progress::progress_bar$new(
format = "[:bar] :percent :pkg",
total = n)
row_dfs <- vector("list", n)
for(i in seq(nrow(pkg_data_names))) {
pkg <- pkg_data_names$package[i]
data_name <- pkg_data_names$data_name[i]
datasets <- pkg_data_names$dataset[[i]]
pb$tick(tokens = list(pkg = format(pkg, width = 12)))
sink(file_, type = "message")
data(list=data_name, package = pkg, envir = env)
row_dfs_i <- lapply(datasets, function(dataset) {
dat <- get(dataset, envir = env)
if(!is.null(filter) && !filter(dat)) return(NULL)
cl <- class(dat)
nms <- names(dat)
nc <- ncol(dat)
if (is.null(nc)) nc <- NA
nr <- nrow(dat)
if (is.null(nr)) nr <- NA
out <- data.frame(
package = pkg,
data_name = data_name,
dataset = dataset,
class = I(list(cl)),
class1 = cl[1],
type = typeof(dat),
names = I(list(nms)),
names_collapsed = paste(nms, collapse = "/"),
nrow = nr,
ncol = nc,
length = length(dat))
if("data.frame" %in% cl) {
classes <- lapply(dat, class)
cl_flat <- unlist(classes)
out <- transform(
out,
classes = I(list(classes)),
types = I(list(vapply(dat, typeof, character(1)))),
logical = sum(cl_flat == 'logical'),
integer = sum(cl_flat == 'integer'),
numeric = sum(cl_flat == 'numeric'),
complex = sum(cl_flat == 'complex'),
character = sum(cl_flat == 'character'),
raw = sum(cl_flat == 'raw'),
list = sum(cl_flat == 'list'),
data.frame = sum(cl_flat == 'data.frame'),
factor = sum(cl_flat == 'factor'),
ordered = sum(cl_flat == 'ordered'),
Date = sum(cl_flat == 'Date'),
POSIXt = sum(cl_flat == 'POSIXt'),
POSIXct = sum(cl_flat == 'POSIXct'),
POSIXlt = sum(cl_flat == 'POSIXlt'))
} else {
out <- transform(
out,
nrow = NA,
ncol = NA,
classes = NA,
types = NA,
logical = NA,
integer = NA,
numeric = NA,
complex = NA,
character = NA,
raw = NA,
list = NA,
data.frame = NA,
factor = NA,
ordered = NA,
Date = NA,
POSIXt = NA,
POSIXct = NA,
POSIXlt = NA)
}
if(is.matrix(dat)) {
out$names <- list(colnames(dat))
out$names_collapsed = paste(out$names, collapse = "/")
}
out
})
row_dfs_i <- do.call(rbind, row_dfs_i)
if(!is.null(row_dfs_i)) row_dfs[[i]] <- row_dfs_i
sink(type = "message")
}
df2 <- do.call(rbind, row_dfs)
df <- merge(df, df2)
df
}


