国庆节快乐,中秋节快乐! O(∩_∩)O 哈哈哈~
The new answer:
Hi, today I do some research on this question, after many times of trial and error, I now get a pretty good solution. The result as follow.

The basic steps are as follows:
- Convert to grayscale, threshold, project to axis, so we can find the watch region.
- Convert to Lab-color-space, so the red needle (in the warm color) can be easily distinguished in the
a channel.
- Normalized the
a channels, threshold, and do morphology operation, so we get an binary mask. After these operations, it will make for finding the potential red needle region.
- Find contours in the mask, filter by some properties, then you'll get it.
1. Find the watch region
# https://i.stack.imgur.com/5oOGL.jpg
imgname = "stkdata/5oOGL.jpg"
img = cv2.imread(imgname)
## Threshold in grayscale
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
retval, threshed = cv2.threshold(gray, 180, 255, cv2.THRESH_BINARY )
## Find wathc region by counting the projector
h,w = img.shape[:2]
x = np.sum(threshed, axis=0)
y = np.sum(threshed, axis=1)
yy = np.nonzero(y>(w/5*255))[0]
xx = np.nonzero(x > (h/5*255))[0]
region = img[yy[0]:yy[-1], xx[0]:xx[-1]]
#cv2.imwrite("region.png", region)
2. Convert to LAB
## Change to LAB space
lab = cv2.cvtColor(region, cv2.COLOR_BGR2LAB)
l,a,b = cv2.split(lab)
imglab = np.hstack((l,a,b))
cv2.imwrite("region_lab.png", imglab)
In the lab-color-space, the red needle can be easily be distinguished.
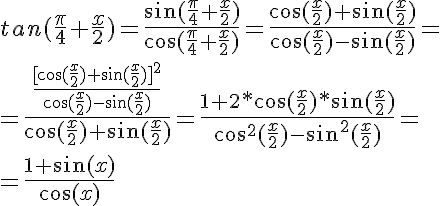
3. Normalized the a channels, threshold, and do the morphology operation.
## normalized the a channel to all dynamic range
na = cv2.normalize(a, None, alpha=0, beta=255, norm_type=cv2.NORM_MINMAX, dtype=cv2.CV_8UC1)
cv2.imwrite("region_a_normalized.png", na)
## Threshold to binary
retval, threshed = cv2.threshold(na, thresh = 180, maxval=255, type=cv2.THRESH_BINARY)
## Do morphology
kernel = cv2.getStructuringElement( cv2.MORPH_ELLIPSE , (3,3))
opened = cv2.morphologyEx(threshed, cv2.MORPH_OPEN,kernel)
res = np.hstack((threshed, opened))
cv2.imwrite("region_a_binary.png", res)
The normalized a channel:
Threshold and do morphology operation, we get a binary mask as the right side.

4. Find contours, and filter them by some properties (such as area and so on).
## Find contours
contours = cv2.findContours(opened, mode=cv2.RETR_LIST, method=cv2.CHAIN_APPROX_SIMPLE)[-2]
## Draw Contours
res = region.copy()
cv2.drawContours(res, contours, -1, (255,0,0), 1)
cv2.imwrite("region_contours.png", res)
## Filter Contours
for idx, contour in enumerate(contours):
bbox = cv2.boundingRect(contour)
area = bbox[-1]*bbox[-2]
if area < 100:
continue
rot_rect = cv2.minAreaRect(contour)
(cx,cy), (w,h), rot_angle = rot_rect
rbox = np.int0(cv2.boxPoints(rot_rect))
cv2.drawContours(res, [rbox], 0, (0,255,0), 1)
text="#{}: {:2.3f}".format(idx, rot_angle)
org=(int(cx)-10,int(cy)-10)
cv2.putText(res, text=text, org = org, fontFace = 1, fontScale=0.8, color=(0,0,255), thickness = 1, lineType=16)
cv2.imwrite("region_result.png", res)
The founded contours:

The result:
We can find that, the #1 contour belong to the red needle, and it's angle is -85.601 .
( The right direction is 0°, the anti-clockwise is negative, and the clockwise is positive)
All Python code is presented here:
#!/usr/bin/python3
# 2017.10.01 22:59:02 CST
# 2017.10.03 23:49:18 CST
import numpy as np
import cv2
# https://i.stack.imgur.com/5oOGL.jpg
imgname = "stkdata/5oOGL.jpg"
img = cv2.imread(imgname)
## Threshold in grayscale
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
retval, threshed = cv2.threshold(gray, 180, 255, cv2.THRESH_BINARY )
## Find wathc region by counting the projector
h,w = img.shape[:2]
x = np.sum(threshed, axis=0)
y = np.sum(threshed, axis=1)
yy = np.nonzero(y>(w/5*255))[0]
xx = np.nonzero(x > (h/5*255))[0]
region = img[yy[0]:yy[-1], xx[0]:xx[-1]]
cv2.imwrite("region.png", region)
## Change to LAB space
lab = cv2.cvtColor(region, cv2.COLOR_BGR2LAB)
l,a,b = cv2.split(lab)
imglab = np.hstack((l,a,b))
cv2.imwrite("region_lab.png", imglab)
## normalized the a channel to all dynamic range
na = cv2.normalize(a, None, alpha=0, beta=255, norm_type=cv2.NORM_MINMAX, dtype=cv2.CV_8UC1)
cv2.imwrite("region_a_normalized.png", na)
## Threshold to binary
retval, threshed = cv2.threshold(na, thresh = 180, maxval=255, type=cv2.THRESH_BINARY)
## Do morphology
kernel = cv2.getStructuringElement( cv2.MORPH_ELLIPSE , (3,3))
opened = cv2.morphologyEx(threshed, cv2.MORPH_OPEN,kernel)
res = np.hstack((threshed, opened))
cv2.imwrite("region_a_binary.png", res)
## Find contours
contours = cv2.findContours(opened, mode=cv2.RETR_LIST, method=cv2.CHAIN_APPROX_SIMPLE)[-2]
## Draw Contours
res = region.copy()
cv2.drawContours(res, contours, -1, (255,0,0), 1)
cv2.imwrite("region_contours.png", res)
## Filter Contours
for idx, contour in enumerate(contours):
bbox = cv2.boundingRect(contour)
area = bbox[-1]*bbox[-2]
if area < 100:
continue
rot_rect = cv2.minAreaRect(contour)
(cx,cy), (w,h), rot_angle = rot_rect
rbox = np.int0(cv2.boxPoints(rot_rect))
cv2.drawContours(res, [rbox], 0, (0,255,0), 1)
text="#{}: {:2.3f}".format(idx, rot_angle)
org=(int(cx)-10,int(cy)-10)
#cv2.putText(res, text=text, org = org, fontFace = cv2.FONT_HERSHEY_PLAIN, fontScale=0.7, color=(0,0,255), thickness = 1, lineType=cv2.LINE_AA)
cv2.putText(res, text=text, org = org, fontFace = 1, fontScale=0.8, color=(0,0,255), thickness = 1, lineType=16)
cv2.imwrite("region_result.png", res)
cv2.imshow("result", res); cv2.waitKey();cv2.destroyAllWindows()
The Origin answer:
I think first you can extract the watch region, then do search red needle in the region, you may change the color space. Here is my sample code in python.
imgname = "5oOGL.jpg"
# https://i.stack.imgur.com/5oOGL.jpg
img = cv2.imread(imgname)
## Threshold in grayscale
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
th, imgbin = cv2.threshold(gray, 180, 255, cv2.THRESH_BINARY )
## Find watch region by projecting and counting
h,w = img.shape[:2]
x = np.sum(imgbin, axis=0)
y = np.sum(imgbin, axis=1)
yy = np.nonzero(y>(w/5*255))[0]
xx = np.nonzero(x > (h/5*255))[0]
region = img[yy[0]:yy[-1], xx[0]:xx[-1]]
cv2.imwrite("region.png", region)
cv2.imshow("region", region); cv2.waitKey(0);# cv2.destroyAllWindows()
## Change to LAB space
lab = cv2.cvtColor(region, cv2.COLOR_BGR2LAB)
l,a,b = cv2.split(lab)
imglab = np.hstack((l,a,b))
cv2.imwrite("imglab.png", imglab)
the watch region

the watch region in lab space.
Some links:
Choosing the correct upper and lower HSV boundaries for color detection with`cv::inRange` (OpenCV)
How to use `cv2.findContours` in different OpenCV versions?
How to find the RED color regions using OpenCV?