I am not an experienced programmer, and needed to fit my data values into a Gaussian graph. The code below was from Gaussian fit for Python.
Using Anaconda, this error message was obtained:
runfile('D:/Anaconda3/gaussian.py', wdir='D:/Anaconda3')
C:\Users\lion\Anaconda3\lib\sitepackages\scipy\optimize\minpack.py:779:OptimizeWarning: Covariance of the parameters could not be estimated category=OptimizeWarning).
The fit gaussian graph was also not as expected; a horizontal curve instead of a gaussian fit curve.
Graph Image:
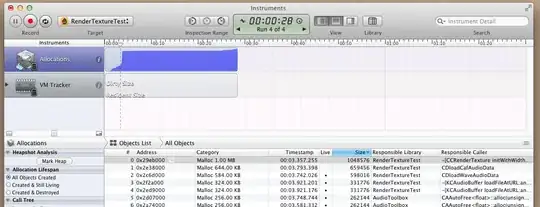
Any help would be appreciated!
Code Used:
import pylab as plb
import matplotlib.pyplot as plt
from scipy.optimize import curve_fit
from scipy import asarray as ar,exp
x = ar(range(399))
y = ar([1, 0, 1, 0, 2, 1, 0, 1, 0, 0, 1, 1, 0, 1, 1, 2, 1, 0, 2, 1, 1, 0, 1, 0, 1, 0, 0, 2, 0, 2,
3, 0, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 1, 0, 1, 1, 1, 1, 2, 0, 3, 3, 0, 1, 2, 1, 1, 2, 3, 1,
4, 1, 1, 1, 0, 1, 0, 3, 4, 0, 1, 3, 0, 2, 0, 3, 0, 0, 0, 3, 2, 0, 2, 0, 0, 1, 2, 0, 0, 0,
3, 2, 1, 0, 1, 3, 4, 2, 4, 1, 2, 1, 1, 2, 0, 2, 2, 6, 2, 4, 2, 0, 1, 2, 2, 3, 4, 6, 2, 3,
2, 4, 1, 4, 9, 6, 6, 4, 5, 6, 3, 7, 8, 9, 7, 8, 8, 4, 10, 10, 12, 9, 18, 18, 16, 14, 13, 13,
15, 17, 16, 26, 24, 37, 34, 36, 40, 48, 52, 52, 50, 56, 68, 90, 71, 107, 93, 117, 134, 207,
200, 227, 284, 287, 337, 379, 449, 471, 626, 723, 848, 954, 1084, 1296, 1481, 1676, 1898, 2024,
2325, 2692, 3110, 3384, 3762, 4215, 4559, 5048, 5655, 6092, 6566, 6936, 7513, 8052, 8414, 9016,
9303, 9598, 9775, 10100, 10265, 10651, 10614, 10755, 10439, 10704, 10233, 10086, 9696, 9467, 9156,
8525, 8200, 7609, 7156, 6678, 6160, 5638, 5227, 4574, 4265, 3842, 3380, 3029, 2767, 2512, 2018, 1856, 1645,
1463, 1253, 1076, 943, 787, 711, 588, 512, 448, 361, 304, 303, 251, 190, 185, 154, 134, 114, 105,
86, 88, 83, 79, 50, 60, 49, 28, 33, 37, 28, 31, 22,
14, 26, 19, 17, 15, 9, 17, 13, 11, 11, 12, 18, 8, 6, 9, 6, 3, 6, 6, 6, 6, 11, 9, 15,
3, 3, 1, 2, 3, 2, 6, 3, 4, 3, 4, 5, 3, 1, 1, 2, 1, 0, 4, 3, 2, 3, 1, 3, 3, 4, 0, 3, 5, 0,
3, 1, 2, 0, 2, 2, 1, 0, 5, 1, 3, 0, 0, 3, 0, 1, 3, 0, 1, 0, 2, 4, 0, 0, 0, 0, 0, 1, 0, 2,
0, 1, 1, 0, 1, 0, 0, 4, 0, 0, 0, 2, 0, 3, 0, 2, 1, 2, 2, 0, 0, 0, 1, 4, 1, 0, 1, 2, 0, 1,
1, 1, 1, 2, 1, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1, 1])
n = len(x) #the number of data
mean = sum(x*y)/n #note this correction
sigma = sum(y*(x-mean)**2)/n #note this correction
def gaus(x,a,x0,sigma):
return a*exp(-(x-x0)**2/(2*sigma**2))
popt,pcov = curve_fit(gaus,x,y,p0=[1,mean,sigma])
plt.plot(x,y,'b+:',label='data')
plt.plot(x,gaus(x,*popt),'ro:',label='fit')
plt.legend()
plt.title('Fig. 3 - Fit for Time Constant')
plt.xlabel('Time (s)')
plt.ylabel('Intensity (Counts)')
plt.show()