I have this data which have outlier . How can i find Mahalanobis disantance and use it to remove outlier.
-
2Mahalanobis distance works for iid data (see [this post for outlier detection](http://kldavenport.com/mahalanobis-distance-and-outliers/)). But your data is not iid. – Maxim Oct 19 '17 at 13:53
3 Answers
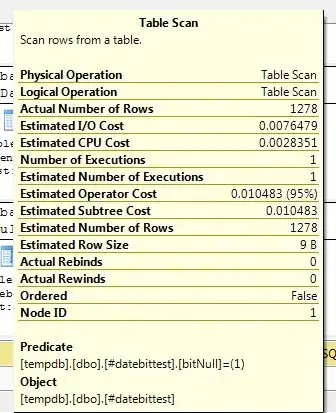
Let me first put some general guidelines:
- Practically speaking, if you have a lot of features and lesser samples, Mahalanobis algorithm tends to give misleading results (you can try it yourself), so the more features you have, the more samples you should provide.
- The covariance matrix must be Symmetric and Positive Definite to make the algorithm works, so you should check before proceeding.
As it's already mentioned, Euclidean Metric fails to find the correct distance because it tries to get ordinary straight-line distance. Thus, if we have multi-dimensional space of variables, two points may look to have the same distance from the Mean, yet one of them is far away from the data cloud (i.e. it's an outlier).
The solution is Mahalanobis Distance which makes something similar to the feature scaling via taking the Eigenvectors of the variables instead of the original axis.
It applies the following formula:
where:
xis the observation to find its distance;mis the mean of the observations;Sis the Covariance Matrix.
Refresher:
The Covariance represents the direction of the relationship between two variables (i.e. positive, negative or zero), so it shows the strength of how one variable is related to the changes of the others.
Implementation
Consider this 6x3 dataset, in which each row represents a sample, and each column represents a feature of the given sample:
First, we need to create a Covariance Matrix of the features of each sample, and that's why we set the parameter rowvar to False in the numpy.cov function, so each column now represents a variable:
covariance_matrix = np.cov(data, rowvar=False)
# data here looks similar to the above table
# in the picture
Next, we find the Inverse of the Covariance Matrix:
inv_covariance_matrix = np.linalg.inv(covariance_matrix)
But before proceeding, we should check, as mentioned above, if the matrix and its inverse are Symmetric and Positive Definite. We use for this Cholesky Decomposition Algorithm, which, fortunately, is already implemented in numpy.linalg.cholesky:
def is_pos_def(A):
if np.allclose(A, A.T):
try:
np.linalg.cholesky(A)
return True
except np.linalg.LinAlgError:
return False
else:
return False
Then, we find the mean m of the variables on each feature (shall I say dimension) and save them in an array like this:
vars_mean = []
for i in range(data.shape[0]):
vars_mean.append(list(data.mean(axis=0)))
# axis=0 means each column in the 2D array
Note that I repeated each row just to avail of matrix subtraction as will be shown next.
Next, we find x - m (i.e. the differential), but since we already have the vectorized vars_mean, all we need to do is:
diff = data - vars_mean
# here we subtract the mean of feature
# from each feature of each example
Finally, apply the formula like this:
md = []
for i in range(len(diff)):
md.append(np.sqrt(diff[i].dot(inv_covariance_matrix).dot(diff[i])))
Note the followings:
- The dimension of the inverse of the covariance matrix is:
number_of_features x number_of_features - The dimension of the
diffmatrix is similar to the original data matrix:number_of_examples x number_of_features - Thus, each
diff[i](i.e. row) is1 x number_of_features. - So according to the Matrix Multiplication rule, the resulted matrix from
diff[i].dot(inv_covariance_matrix)will be1 x number_of_features; and when we multiply again bydiff[i];numpyautomatically considers the latter as a column matrix (i.e.number_of_features x 1); so the final result will become a single value (i.e. no need for transpose).
In order to detect outliers, we should specify a threshold; but since the square of Mahalanobis Distances follow a Chi-square distribution with a degree of freedom = number of feature in the dataset, then we can choose a threshold of say 0.1, then we can use chi2.cdf method from Scipy, like this:
1 - chi2.cdf(square_of_mahalanobis_distances, degree_of_freedom)
So any point that has (1 - chi-squared CDF) that less than or equal the threshold, can be classified as an outlier.
Putting All Together
import numpy as np
def create_data(examples=50, features=5, upper_bound=10, outliers_fraction=0.1, extreme=False):
'''
This method for testing (i.e. to generate a 2D array of data)
'''
data = []
magnitude = 4 if extreme else 3
for i in range(examples):
if (examples - i) <= round((float(examples) * outliers_fraction)):
data.append(np.random.poisson(upper_bound ** magnitude, features).tolist())
else:
data.append(np.random.poisson(upper_bound, features).tolist())
return np.array(data)
def MahalanobisDist(data, verbose=False):
covariance_matrix = np.cov(data, rowvar=False)
if is_pos_def(covariance_matrix):
inv_covariance_matrix = np.linalg.inv(covariance_matrix)
if is_pos_def(inv_covariance_matrix):
vars_mean = []
for i in range(data.shape[0]):
vars_mean.append(list(data.mean(axis=0)))
diff = data - vars_mean
md = []
for i in range(len(diff)):
md.append(np.sqrt(diff[i].dot(inv_covariance_matrix).dot(diff[i])))
if verbose:
print("Covariance Matrix:\n {}\n".format(covariance_matrix))
print("Inverse of Covariance Matrix:\n {}\n".format(inv_covariance_matrix))
print("Variables Mean Vector:\n {}\n".format(vars_mean))
print("Variables - Variables Mean Vector:\n {}\n".format(diff))
print("Mahalanobis Distance:\n {}\n".format(md))
return md
else:
print("Error: Inverse of Covariance Matrix is not positive definite!")
else:
print("Error: Covariance Matrix is not positive definite!")
def is_pos_def(A):
if np.allclose(A, A.T):
try:
np.linalg.cholesky(A)
return True
except np.linalg.LinAlgError:
return False
else:
return False
data = create_data(15, 3, 10, 0.1)
print("data:\n {}\n".format(data))
MahalanobisDist(data, verbose=True)
Result
data:
[[ 12 7 9]
[ 9 16 7]
[ 14 11 10]
[ 14 5 5]
[ 12 8 7]
[ 8 8 10]
[ 9 14 8]
[ 12 12 10]
[ 18 10 6]
[ 6 12 11]
[ 4 12 15]
[ 5 13 10]
[ 8 9 8]
[106 116 97]
[ 90 116 114]]
Covariance Matrix:
[[ 980.17142857 1143.62857143 1035.6 ]
[1143.62857143 1385.11428571 1263.12857143]
[1035.6 1263.12857143 1170.74285714]]
Inverse of Covariance Matrix:
[[ 0.03021777 -0.03563241 0.0117146 ]
[-0.03563241 0.08684092 -0.06217448]
[ 0.0117146 -0.06217448 0.05757261]]
Variables Mean Vector:
[[21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8], [21.8, 24.6, 21.8]]
Variables - Variables Mean Vector:
[[ -9.8 -17.6 -12.8]
[-12.8 -8.6 -14.8]
[ -7.8 -13.6 -11.8]
[ -7.8 -19.6 -16.8]
[ -9.8 -16.6 -14.8]
[-13.8 -16.6 -11.8]
[-12.8 -10.6 -13.8]
[ -9.8 -12.6 -11.8]
[ -3.8 -14.6 -15.8]
[-15.8 -12.6 -10.8]
[-17.8 -12.6 -6.8]
[-16.8 -11.6 -11.8]
[-13.8 -15.6 -13.8]
[ 84.2 91.4 75.2]
[ 68.2 91.4 92.2]]
Mahalanobis Distance:
[1.3669401667524865, 2.1796331318432967, 0.7470525416547134, 1.6364973119931507, 0.8351423113609481, 0.9128858131134882, 1.397144258271586, 0.35603382066414996, 1.4449501739129382, 0.9668775289588046, 1.490503433100514, 1.4021488309805878, 0.4500345257064412, 3.239353067840299, 3.260149280200771]
- 13,349
- 6
- 30
- 42
-
3Minimal? Wow. Impressed. It looks like you put quite some work into this. Lets hope that many people recognize the effort. – GhostCat Aug 17 '18 at 11:47
-
Thanks. This was extremely helpful. A question: what do you do if your matrix is not positive semidefinite or symmetric? – Code Pope Mar 06 '19 at 19:03
-
Your code has also a problem. You are using the variable `points` in the `MD_detectOutliers` function which is not defined anywhere. I could not understand what `points` are because you are computing the mean of them (so I suggest that they are vectors from R^n) but you compare them with scalars: `MD[i] > up_t`? I mean `MD[i]` is a scalar, but as I know `up_t` is a vector. How should this work? – Code Pope Mar 06 '19 at 19:22
-
But now as I run your code, no outlier is detected although there do exist outliers in the `data`. Maybe you should check the code again. – Code Pope Mar 06 '19 at 19:34
-
@CodePope For `MD` in particular, it just fails to detect outliers if it is not positive semidefinite or symmetric. However, there is a very popular algorithm called Minimum Covariance Determinant [MCD](https://scikit-learn.org/stable/modules/generated/sklearn.covariance.EllipticEnvelope.html#sklearn.covariance.EllipticEnvelope) that uses Mahalanobis Distances in a better approach. Continue in another comment.. – Yahya Mar 06 '19 at 19:36
-
@CodePop look [here](https://github.com/scikit-learn/scikit-learn/blob/7b136e92acf49d46251479b75c88cba632de1937/sklearn/covariance/robust_covariance.py) and you can see how they check for the dimension (i.e. number of features) and how they treat it. And [here](https://wis.kuleuven.be/stat/robust/papers/2010/wire-mcd.pdf) is the algorithm paper. – Yahya Mar 06 '19 at 19:36
-
1@CodePope Thanks very much for pointing out that issue. `MD` should be plugged in instead of `data`. I fixed it and it's working now for sure. Sorry for any inconvenience and Thanks again. – Yahya Mar 06 '19 at 19:47
-
I have a doubt. The last part of the code assumes that the mahalanobis distance follows a normal distribution and hence uses the 68–95–99.7 rule. But it is to be noted that MD follows a chi square distribution. – user3284005 Feb 07 '21 at 10:10
-
As @Yahya pointed out earlier , in order to calculate the MD the covariance matrix should be positive and semi definite . These are two necessary conditions in order to take the inverse of the covariance matrix . The inverse of the matrix is calculated – Gaurav Chawla Jan 11 '22 at 03:09
-
Thanks for your comprehensive answer. It was very helpful. I am going to use Mahalanobis Distance to detect anomalies, but my covariance matrix is not positive definite (it is symmetric) (The last eigenvector is zero). What should I do now? – 8Simon8 Jul 05 '22 at 19:49
In multivariate data, Euclidean distance fails if there exists covariance between variables (i.e. in your case X, Y, Z). 
Therefore, what Mahalanobis Distance does is,
It transforms the variables into uncorrelated space.
Make each variables varience equals to 1.
Then calculate the simple Euclidean distance.
We can calculate the Mahalanobis Distance for each data sample as follows,
Here, I have provided the python code and added the comments so that you can understand the code.
import numpy as np
data= np.matrix([[1, 2, 3, 4, 5, 6, 7, 8],[1, 4, 9, 16, 25, 36, 49, 64],[1, 4, 9, 16, 25, 16, 49, 64]])
def MahalanobisDist(data):
covariance_xyz = np.cov(data) # calculate the covarince matrix
inv_covariance_xyz = np.linalg.inv(covariance_xyz) #take the inverse of the covarince matrix
xyz_mean = np.mean(data[0]),np.mean(data[1]),np.mean(data[2])
x_diff = np.array([x_i - xyz_mean[0] for x_i in x]) # take the diffrence between the mean of X variable the sample
y_diff = np.array([y_i - xyz_mean[1] for y_i in y]) # take the diffrence between the mean of Y variable the sample
z_diff = np.array([z_i - xyz_mean[2] for z_i in z]) # take the diffrence between the mean of Z variable the sample
diff_xyz = np.transpose([x_diff, y_diff, z_diff])
md = []
for i in range(len(diff_xyz)):
md.append(np.sqrt(np.dot(np.dot(np.transpose(diff_xyz[i]),inv_covariance_xyz),diff_xyz[i]))) #calculate the Mahalanobis Distance for each data sample
return md
def MD_removeOutliers(data):
MD = MahalanobisDist(data)
threshold = np.mean(MD) * 1.5 # adjust 1.5 accordingly
outliers = []
for i in range(len(MD)):
if MD[i] > threshold:
outliers.append(i) # index of the outlier
return np.array(outliers)
print(MD_removeOutliers(data))
Hope this helps.
References,
- 1,839
- 11
- 13
-
I can't find the library having MahalanobisDist please tell the library.And it will be helpful if you explain it . – Shubham Tyagi Oct 20 '17 at 14:55
-
awesome answer! Now can you tell me why openCv's Mahalanobis asks for multiple sets of data? (data1,data2,inverted_covariance) – john k Nov 19 '17 at 20:30
-
As @Yahya pointed out earlier, in order to calculate the MD the cov .matrix should be positive and semi-definite. These are necessary conditions in order to take the inverse of the cov. matrix . The inverse of the matrix uses the reciprocal of the determinant of the matrix. The fact that you may be unable to calculate the determinant for the matrix may point to deeper problem in the data set itself. It can be because two or more columns in the dataset are correlated . You would be better off removing such pairs before trying to calculate MD. Using Pseudo Inverse can be another alternative.
- 368
- 4
- 6