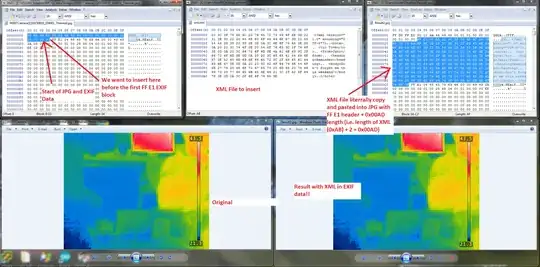
I have a vector of means and standard deviations, and I would like to plot the densities corresponding to these means and standard deviations in the same plot using ggplot2. I used mapply and gather to solve this problem, but it's quite a lot of lines of code for something which I think should be trivial:
library(dplyr)
library(tidyr)
library(ggplot2)
# generate data
my_data <- data.frame(mean = c(0.032, 0.04, 0.038, 0.113, 0.105, 0.111),
stdev = c(0.009, 0.01, 0.01, 0.005, 0.014, 0.006),
test = factor(c("Case_01", "Case_02", "Case_03", "Case_04",
"Case_05", "Case_06")))
# points at which to evaluate the Gaussian densities
x <- seq(-0.05, 0.2, by = 0.001)
# build list of Gaussian density vectors based on means and standard deviations
pdfs <- mapply(dnorm, mean = my_data$mean, sd = my_data$stdev, MoreArgs = list(x = x),
SIMPLIFY = FALSE)
# add group names
names(pdfs) <- my_data$test
# convert list to dataframe
pdfs <- do.call(cbind.data.frame, pdfs)
pdfs$x <- x
# convert dataframe to tall format
tall_df <- gather(pdfs, test, density, -x)
# build plot
p <- ggplot(tall_df, aes(color = test, x = x, y = density)) +
geom_line() +
geom_segment(data = my_data, aes(color = test, x = mean, y = 0,
xend = mean, yend = 100), linetype = "dashed") +
coord_cartesian(ylim = c(-1, 100))
print(p)
Plot multiple normal curves in same plot
and as a matter of fact, the accepted answer uses mapply, so that confirms me that I'm on the right track. However, what I don't like of that answer is that it hard-codes means and standard deviations in the mapply call. This won't work in my use case, because I read the real data from disk (of course, in the MRE I skipped the data reading part for simplicity). Is it possible to simplify my code, without sacrificing readability, and without hard-coding the mean and standard deviation vectors in the mapply call?
EDIT maybe the call to mapply may be avoided by using the package mvtnorm, but I don't think that affords any real simplification here. Most of my code comes after the call to mapply.