I have an array of subplots, but whenever I try to reduce the tick density or even change the format, the values seem to change.
- Is there a distinction here between the 'tick' and its value? How would I match the correct value to the tick if so?
Here's a stand-alone plot that's right: France
Here's the (EDITED) code I'm trying:
import matplotlib as mpl
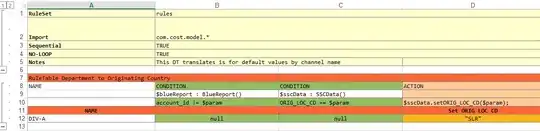
from matplotlib.ticker import FormatStrFormatter
f, axarr = plt.subplots(5,6,figsize=(30,30))
for i in range(0,6):
for j in range(0,5):
# there are only 26 entries in ser
if (4*i+j) < 26:
x = pp_gdp.loc[pp_gdp.index == ser.index[4*i+j],:].round(1)
y = ser.loc[ser.index == ser.index[4*i+j],:].round(1)
axarr[j,i].scatter(x,y)
axarr[j,i].set_title(ser.index[4*i+j])
xt = axarr[j,i].get_xticks()
#axarr[j,i].set_xscale('linear')
axarr[j,i].set_xticks([axarr[j,i].get_xticks()[0],int(len(axarr[j,i].get_xticks())/2), axarr[j,i].get_xticks()[-1]])
axarr[j,i].set_xticklabels([x.values.min(),sorted(x.values)[0][int(len(xt)/2)],x.values.max()])
yt = axarr[j,i].get_yticks()
#axarr[j,i].set_yscale('linear')
axarr[j,i].set_yticks([axarr[j,i].get_yticks()[0],int(len(axarr[j,i].get_yticks())/2), axarr[j,i].get_yticks()[-1]])
axarr[j,i].set_yticklabels([y.values.min(),sorted(y.values)[0][int(len(yt)/2)],y.values.max()]);
Here's what I get if I comment out all the tick code, i.e. the default subplots: Default plot
As you can see, it's far too dense to read.
Here's what happens when I try nbins:
axarr[j,i].locator_params(tight=True, axis='y', nbins=10)
axarr[j,i].locator_params(tight=True, axis='x', nbins=5)
Again, the values are wrong on both axes.
Using the get_xticks and set_xticks as in the big code sample sort of works, but ruins the intuition of the plot. Also the scale is all wrong because of the way it keeps the 0.
Very linear plots stay linear, but the slope is exaggerated. Plots a little more complicated lose their shape altogether, as in: China now wrong
The chart on the rightis much harder to understand, just because the scale isn't playing nicely.
With my new
Thanks, Roy