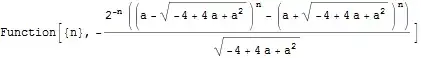I am writing a Shiny App based on Cointegration.
After I find the cointegration vectors I plot all the mean reverting series using ggplot2. The way I do it is I put all these cointegration series in a dataframe and I melt it then I facet wrap based on the variable.
Until here I had no problem.
Now I want to add the moving average and the moving standard deviation to the plot of each cointegration series but I can't find a way to have each series with it's MA and STD alone.
I know the problem with what I have right now is with the way I am facet_wraping but I am not an experienced user so I do not know how to solve it. Now I have:
I want the Cointegration Series 1, MA series1 and Std Series 1 to be on the same graph and have different colours and the same for Cointegration Series 2.... The code I use is:
m= melt(DF, id.vars = "Date") # melt the df
ggplot(m, aes(x = Date, y = value, colour = variable, group=variable)) +geom_line() + facet_wrap( ~ variable) # plot
Is there a way to wrap the first 3 columns together ? Should I melt in a different way ?? Do I need to explain more to be more clear ?
EDIT: Basically in Lehman's terms suppose I have a dataframe df composed of the columns:
Date, S1, MA1, STD1, S2, MA2, STD2
I want my x axis to be Date, and I want to have S1, MA1, STD1 on a plot and S2, MA2 and STD2 on another plot using ggplot2. As id I were facet wraping S1,MA1, STD1 together and then S2, MA2, STD2 also together
Suppose this is the DataFrame:
Date S1 M1 ST1 S2 M2 ST2
1 02/12/1999 -0.000217052 0.002862195 -0.002390842 2.2025825 -1.17258213 -0.3057015
2 03/12/1999 -0.004882038 -0.015920939 -0.007014382 -0.4040079 -0.69496488 0.1000167
3 06/12/1999 -0.001445954 0.005077610 0.000000000 1.1573779 -1.64268166 -0.3847015
4 07/12/1999 -0.000907952 -0.005403168 0.002027728 -0.3165827 -1.16466940 -1.0113501
5 08/12/1999 0.000881220 0.011375226 -0.013524439 0.5154455 -0.51725208 1.0942177
6 09/12/1999 0.001043752 0.013400502 0.017033342 1.9368608 -0.05587143 0.5203337
Is it clear enough now ?