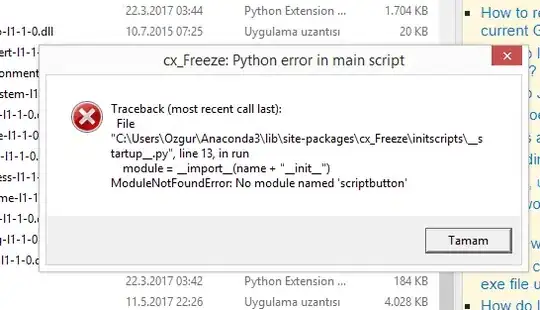
This is somewhat tricky with ggplot, so please forgive the long, convoluted, and admittedly a bit hacky answer. The basic problem is that with coord_fixed, the height of the y-axis becomes inextricably linked to the length of the x-axis.
There are two ways we can break this dependency:
by using the expand argument of scale_y_continuous. This allows us to extend the y axis by a given amount beyond the range of the data. The tricky bit is knowing how much to expand it, because this depends in a hard-to-predict way on all elements of the plot, including how many facets there are and the size of axis titles and labels etc.
by allowing the width of the two plots to differ. The tricky thing here is, as above, how to find the correct width as this depends on the various other aspects of the plots.
First I show how we can solve the first version (how much to expand the y-axis). Then using a similar approach and a little extra trickery we can also solve the varying width version.
Solution to finding how much to expand the y-axis
Given the difficulties of predicting how large the plotting area will be (which depnds on the relative sizes of all the elements of the plot), what we can do is to save a dummy plot in which we shade the plot area in black, read the image file back in, then measure the size of the black area to determine how large the plot area is:
1) let's start by assigning your plots to variables
p1 = ggplot(df1) +
geom_point(aes(value1, value2)) +
facet_grid(~var1) +
coord_fixed()
p2 = ggplot(df1) +
geom_point(aes(value1, value2)) +
facet_grid(~var2) +
coord_fixed()
2) now we can save some dummy versions of these plots that only show a black rectangle where the plotting region is:
t_blank = theme(strip.background = element_rect(fill = NA),
strip.text = element_text(color=NA),
axis.title = element_text(color = NA),
axis.text = element_text(color = NA),
axis.ticks = element_line(color = NA))
p1 + geom_rect(aes(xmin=-Inf, xmax=Inf, ymin=-Inf, ymax=Inf), fill='black') +
t_blank
ggsave(fn1 <- tempfile(fileext = '.png'), height=5, units = 'in')
p2 + geom_rect(aes(xmin=-Inf, xmax=Inf, ymin=-Inf, ymax=Inf), fill='black') +
t_blank
ggsave(fn2 <- tempfile(fileext = '.png'), height=5, units = 'in')
3) then we read these into an array (just the first color band is enough)
library(png)
p1.saved = readPNG(fn1)[,,1]
p2.saved = readPNG(fn2)[,,1]
4) calculate the height of each plotting area (the black-shaded areas which have a value=zero)
p1.height = diff(row(p1.saved)[range(which(p1.saved==0))])
p2.height = diff(row(p2.saved)[range(which(p2.saved==0))])
5) Find how much we need to expand the plotting area based on these. Note that we subtract the ratio of heights from 1.1 to account for the fact that the original plots were already expanded by the default amount of 0.05 in each direction. Disclaimer -- this formula works on your example. I haven't had time to check it more broadly, and it may yet need adapting to ensure generality for other plots
height.expand = 1.1 - p2.height / p1.height
6) Now we can save the plots using this expansion factor
ggsave("plot_2facet.pdf", p1, height=5, units = 'in')
ggsave("plot_4facet.pdf", p2 + scale_y_continuous(expand=c(height.expand, 0)),
height=5, units = 'in')
Solution to finding how much to alter the width
first, lets set the width of the first plot to what we want
p1.width = 10
Now, using the same approach as in the previous section we find how tall the plotting area is in this plot.
p1 + geom_rect(aes(xmin=-Inf, xmax=Inf, ymin=-Inf, ymax=Inf), fill='black') +
t_blank
ggsave(fn1 <- tempfile(fileext = '.png'), height=5, width = p1.width, units = 'in')
p1.saved = readPNG(fn1, info = T)[,,1]
p1.height = diff(row(p1.saved)[range(which(p1.saved==0))])
Next, we find the mimimum width the second plot must have to get the same height (note - we look for a minimum here because any greater width than this will not increase the height, which already fiulls the vertical space, but will simply add white space to the left and right)
We will solve for the width using the function uniroot which finds where a function crosses zero. To use uniroot we first define a function that will calculate the height of a plot given its width as an argument. It then returns the difference between that height and the height we want. The line if (x==0) x = -1e-8 in this function is a dirty trick to allow uniroot solve a function that reaches zero, but does not cross it - see here.
fn2 <- tempfile(fileext = '.png')
find.p2 = function(w){
p = p2 + geom_rect(aes(xmin=-Inf, xmax=Inf, ymin=-Inf, ymax=Inf), fill='black') +
t_blank
ggsave(fn2, p, height=5, width = w, units = 'in')
p2.saved = readPNG(fn2, info = T)[,,1]
p2.height = diff(row(p2.saved)[range(which(p2.saved==0))])
x = abs(p1.height - p2.height)
if (x==0) x = -1e-8
x
}
N1 = length(unique(df$var1))
N2 = length(unique(df$var2))
p2.width = uniroot(find.p2, c(p1.width, p1.width*N2/N1))
Now we are ready to save the plots with the correct widths to ensure they have the same height.
p1
ggsave("plot_2facet.pdf", height=5, width = p1.width, units = 'in')
p2
ggsave("plot_4facet.pdf", height=5, width = p2.width$root, units = 'in')