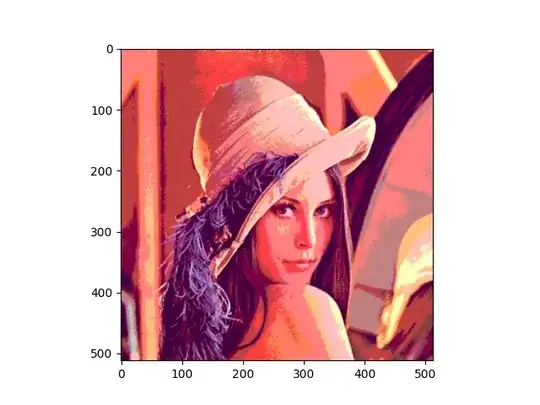
I am trying to plot the parameter estimates and levels of hierarchy from a stan model output. For the legend, I am hoping to remove all labels except for the "Overall Effects" label but I can't figure out how to remove all of the species successfully.
Here is the code:
ggplot(dfwide, aes(x=Estimate, y=var, color=factor(sp), size=factor(rndm),
alpha=factor(rndm))) +
geom_point(position =pd) +
geom_errorbarh(aes(xmin=(`2.5%`), xmax=(`95%`)), position=pd,
size=.5, height = 0, width=0) +
geom_vline(xintercept=0) +
scale_colour_manual(values=c("blue", "red", "orangered1","orangered3", "sienna4",
"sienna2", "green4", "green3", "purple2", "magenta2"),
labels=c("Overall Effects", expression(italic("A. pensylvanicum"),
italic("A. rubrum"), italic("A. saccharum"),
italic("B. alleghaniensis"), italic("B. papyrifera"),
italic("F. grandifolia"), italic("I. mucronata"),
italic("P. grandidentata"), italic("Q. rubra")))) +
scale_size_manual(values=c(3, 1, 1, 1, 1, 1, 1, 1, 1, 1)) +
scale_shape_manual(labels="", values=c("1"=16,"2"=16)) +
scale_alpha_manual(values=c(1, 0.4)) + guides(size=FALSE, alpha=FALSE) +
ggtitle(label = "A.") +
scale_y_discrete(limits = rev(unique(sort(dfwide$var))), labels=estimates) +
ylab("") +
labs(col="Effects") + theme(legend.title=element_blank())