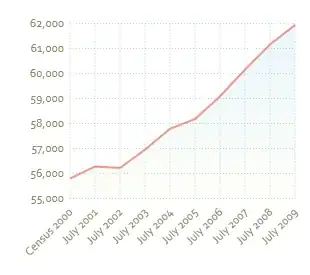
I would like to set the thickness of geom_line to the proportion of data that follows that path, in the same way that geom_count sets the size of points based on the proportion of data that overlap at that point, or find a function that will allow me to do this.
I would also be happy if I could do this as a count rather than a proportion - either would work. I have attached the graph the grey lines represent connections between the same ID (ie. same individual in different categories), if I could set the thickness of the lines I can show the most common connection pathways.
My current code is:
ggplot(dat, aes(x = Category, y = Metric, group = ID)) +
geom_line(aes(group = ID), colour = "gray59") +
geom_count(aes(size = ..prop.., group = 1), colour = "gray59") +
scale_size_area(max_size = 5) +
theme_bw() +
geom_smooth(method = "lm", se = F, colour = "black",
aes(group = 1), linetype = "dotdash") +
xlab("Category") +
ylab("Metric") +
theme(text = element_text(size = 16))
This is the resulting graph, point size shows the proportion of data that overlaps at that point, I would like to do the same with line thickness if possible:
My searching has so far turned up nothing helpful but maybe I am searching the wrong terms. Any help would be much appreciated!
Here is the data - unsure how to upload it as a file
dat <- structure(list(IDD = structure(c(1L, 1L, 1L, 1L, 3L, 3L, 4L,
4L, 4L, 4L, 4L, 5L, 5L, 5L, 6L, 6L, 2L, 2L, 2L, 2L, 7L, 7L, 7L,
8L, 8L, 8L, 9L, 9L, 9L, 9L, 10L, 10L, 10L, 10L, 11L, 11L, 12L,
12L, 13L, 13L, 13L, 13L, 14L, 14L, 15L, 15L, 15L, 15L, 16L, 16L,
16L, 16L, 17L, 17L, 18L, 18L, 18L, 18L, 19L, 19L, 20L, 20L, 21L,
21L, 21L, 22L, 22L, 23L, 23L, 24L, 24L, 25L, 25L, 25L, 26L, 26L,
26L, 26L, 27L, 27L, 28L, 28L, 29L, 29L, 29L, 30L, 30L, 30L, 31L,
31L, 31L, 31L, 32L, 32L, 33L, 33L, 33L, 34L, 34L, 34L, 34L, 35L,
35L, 36L, 36L, 36L, 37L, 37L, 37L, 37L, 38L, 38L, 38L, 39L, 39L,
39L, 40L, 40L, 40L, 41L, 41L, 42L, 42L, 43L, 43L, 44L, 44L, 44L,
44L, 45L, 45L, 45L, 46L, 46L, 46L, 47L, 47L, 47L, 48L, 48L, 49L,
49L, 50L, 50L, 51L, 51L, 51L, 51L, 52L, 52L, 53L, 53L, 54L, 54L,
55L, 55L, 56L, 56L, 57L, 57L, 57L, 58L, 58L, 59L, 59L, 59L, 59L
), .Label = c("ID005", "ID040", "ID128", "ID131", "ID133", "ID134",
"ID147", "ID149", "ID166", "ID167", "ID175", "ID181", "ID191",
"ID198", "ID213", "ID235", "ID254", "ID257", "ID259", "ID273",
"ID279", "ID287", "ID292", "ID299", "ID300", "ID321", "ID334",
"ID348", "ID349", "ID354", "ID359", "ID377", "ID379", "ID383",
"ID390", "ID395", "ID409", "ID445", "ID467", "ID469", "ID482",
"ID492", "ID496", "ID524", "ID526", "ID527", "ID534", "ID535",
"ID538", "ID545", "ID564", "ID576", "ID578", "ID579", "ID600",
"ID610", "ID622", "ID631", "ID728"), class = "factor"), Category = c(2L,
4L, 5L, 5L, 2L, 4L, 1L, 3L, 3L, 4L, 4L, 2L, 4L, 5L, 5L, 5L, 2L,
5L, 5L, 5L, 3L, 2L, 5L, 4L, 5L, 5L, 4L, 4L, 5L, 5L, 3L, 4L, 5L,
5L, 2L, 4L, 2L, 5L, 3L, 4L, 5L, 5L, 4L, 5L, 3L, 4L, 5L, 5L, 3L,
4L, 5L, 5L, 5L, 5L, 2L, 3L, 4L, 4L, 5L, 5L, 5L, 5L, 4L, 4L, 5L,
5L, 5L, 3L, 4L, 5L, 5L, 4L, 5L, 5L, 1L, 3L, 4L, 4L, 3L, 5L, 3L,
5L, 2L, 3L, 4L, 3L, 4L, 4L, 3L, 3L, 4L, 4L, 3L, 5L, 3L, 4L, 4L,
3L, 3L, 4L, 5L, 2L, 3L, 2L, 3L, 4L, 2L, 2L, 3L, 4L, 4L, 5L, 5L,
2L, 3L, 4L, 2L, 3L, 4L, 3L, 4L, 4L, 5L, 3L, 4L, 1L, 2L, 3L, 4L,
1L, 3L, 4L, 1L, 3L, 4L, 1L, 3L, 4L, 3L, 4L, 3L, 3L, 2L, 3L, 2L,
2L, 3L, 3L, 2L, 3L, 2L, 3L, 3L, 4L, 3L, 4L, 3L, 4L, 1L, 2L, 3L,
2L, 3L, 1L, 3L, 4L, 4L), Metric = c(2, 2, 3.5, 4, 2, 1.5, 2,
2, 3, 3, 2, 2, 2, 2, 3.5, 3.5, 2, 3, 3.5, 4, 2, 2, 3, 2, 3, 3,
2, 3, 3, 2.5, 1.5, 3, 3.5, 4, 2, 2, 1.5, 2, 1.5, 2, 2, 2, 2.5,
3, 2.5, 3.5, 3.5, 3.5, 1.5, 2, 2.5, 2.5, 3.5, 4, 2, 2, 1.5, 3,
3.5, 3, 3, 3, 3.5, 2.5, 3, 3, 3, 2, 3, 2.5, 2.5, 2, 2, 2, 2,
2, 2, 2, 2.5, 2.5, 2, 3, 2.5, 2, 2.5, 2, 2.5, 2.5, 2, 2, 2.5,
3.5, 2, 2.5, 2.5, 2.5, 2.5, 2, 2, 2, 2.5, 2, 2, 1.5, 2, 2, 2.5,
2, 2, 2.5, 2, 2, 2.5, 2.5, 2.5, 3, 2.5, 2.5, 2.5, 2, 2, 2.5,
2.5, 2, 2, 2, 2, 1.5, 2, 1.5, 2, 2, 2, 1.5, 2, 2, 2.5, 2.5, 1.5,
1.5, 2, 2.5, 2, 2, 2, 2, 2.5, 2, 1.5, 2, 2.5, 2, 1.5, 1.5, 1.5,
2, 2, 2, 2, 2, 1.5, 2, 2.5, 2, 2, 2.5, 2.5)), .Names = c("IDD",
"Category", "Metric"), class = "data.frame", row.names = c(NA,
-167L))