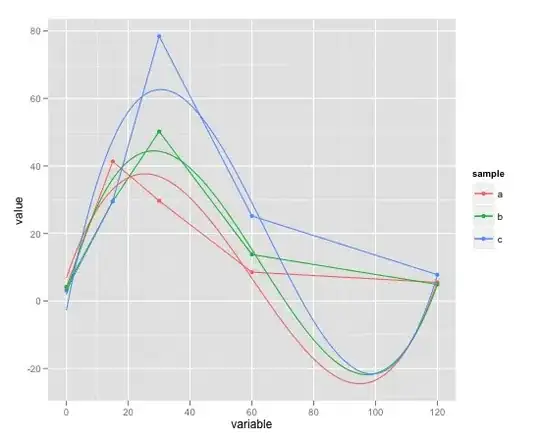
I have two plots that share the y-axis but have different x-axes. I used facet_grid to separate them appropriately (see figure), but the two x-axes need to have different titles (instead of the one title "Num Cell Lines.Tissue"). I've seen something similar done by creating 2 ggplot objects first then use a function like ggplotGrob. Is there a way to do this directly?
Edit 1: to include data, code
Data:
Variable Condition Num.CellLines.Tissue Percent.Altered
V1 C1 1 0.20149254
V1 C1 2 0.03731343
V1 C1 3 0
V1 C1 4 0
V1 C1 5 0
V2 C2 1 0.74893617
V2 C2 2 0.37446809
V2 C2 3 0.16595745
V2 C2 4 0.09787234
V2 C2 5 0.06808511
V2 C2 6 0.05531915
V2 C2 7 0.02553191
V2 C2 8 0.01702128
V2 C2 9 0.01276596
V2 C2 10 0.00851064
V2 C3 1 0.88554217
V2 C3 2 0.68072289
V2 C3 3 0.40361446
V2 C3 4 0.22289157
V2 C3 5 0.11445783
V2 C3 6 0.06626506
V2 C3 7 0.04819277
V2 C3 8 0.01807229
V2 C3 9 0.01807229
V2 C3 10 0.01204819
V2 C4 1 0.87301587
V2 C4 2 0.6984127
V2 C4 3 0.52380952
V2 C4 4 0.38095238
V2 C4 5 0.25925926
V2 C4 6 0.14285714
V2 C4 7 0.07407407
V2 C4 8 0.04232804
V2 C4 9 0.03703704
V2 C4 10 0.03174603
Code:
ggplot(data, aes(y=Percent.Altered, x = Num.CellLines.Tissue, color= Condition )) + geom_line(size=1) + facet_grid(. ~ Variable, scales="free_x")
Edit 2: Image of ideal plot
The labels V1 and V2 show up the way I want them to by default, and they are different from what the titles of the x-axes below them should be.