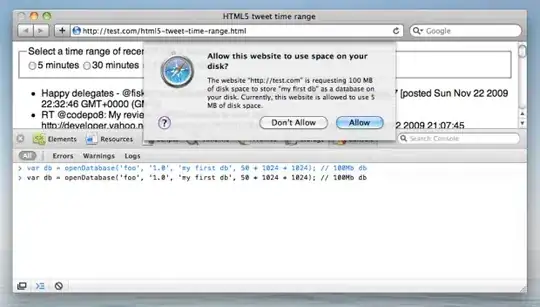
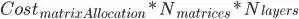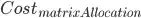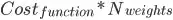I'm trying to generalize a neural network function to arbitrarily many layers, and so I need multiple matrices to hold the weights for each neuron in each layer. I was originally explicitly declaring matrix objects in R to hold my weights for each layer. Instead of having one matrix per layer, I thought of a way (not saying it's original), to store all of my weights in a single array and defined an "indexing function" to map a weight to its appropriate index in the array.
I defined the function as follows:
where  is the k-th weight of the j-th neuron in the i-th layer and L(r) is the number of neurons in layer r. After writing these definitions, I realize that stackoverflow doesn't allow latex like mathoverflow which is unfortunate.
Now the question is: Is it more efficient to compute the index of my weights in this way, or is actually less efficient?
After looking up how indices are computed for arrays in general, this is essentially what is done on compilation anyway if I just kept a matrix in each layer holding the weights, so it seems like I may just be making my code overly complicated and harder to understand if there's no difference in time efficiency.
is the k-th weight of the j-th neuron in the i-th layer and L(r) is the number of neurons in layer r. After writing these definitions, I realize that stackoverflow doesn't allow latex like mathoverflow which is unfortunate.
Now the question is: Is it more efficient to compute the index of my weights in this way, or is actually less efficient?
After looking up how indices are computed for arrays in general, this is essentially what is done on compilation anyway if I just kept a matrix in each layer holding the weights, so it seems like I may just be making my code overly complicated and harder to understand if there's no difference in time efficiency.