I am trying to learn how to integrate python SymPy with SciPy for numerically solving ordinary differential equations. However, I was kinda lost about how to actually get the SymPy form of a first order system of ODEs into a format that I can process with scipy.integrate.odeint()
Note, some people suggested that this is similar to another post, but that is not the case. The other post is here.
Convert sympy expressions to function of numpy arrays
So this other post is a much more complicated case where the user wants to accelerate computation of an ODE using Theano or some other libraries. I am just trying to understand the basic interface between SymPy and SciPy, so this other post is not at all helpful.
As a toy example, I was using the Lotka-Volterra equation to test the use of SymPy. The equations are:
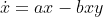
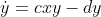
I can solve this in the conventional way with Scipy and it works. Here is the working code.
import numpy as np
import scipy
from scipy.integrate import odeint, ode, solve_ivp
import sympy
import matplotlib.pyplot as plt
sympy.init_printing()
def F_direct(X, t, args):
F = np.zeros(2)
a, b, c, d = args
x,y = X
F[0] = a*x - b*x*y
F[1] = c*x*y- d*y
return F
argst = [0.4,0.002,0.001,0.7]
xy0 = [600, 400]
t = np.linspace(0, 50, 250)
xy_t, infodict = odeint(F_direct, xy0, t, args=(argst,), full_output=True)
plt.plot(t, xy_t[:,1], 'o', t, xy_t[:,0])
plt.grid(True)
plt.xlabel('x'); plt.ylabel('y')
plt.legend(('Numerical', 'Exact'), loc=0)
plt.show()
Now I was kind lost about how to do this with SymPy. I have a sense of what needs to be done, but was not sure how to proceed. The only example I have found is way too complicated to learn from. Here is what I have.
x, y, a, b, c, d = sympy.symbols('x y a b c d')
t = np.linspace(0, 50, 250)
ode1 = sympy.Eq(a*x - b*x*y)
ode2 = sympy.Eq(c*x*y - d*y)
I am supposed to put these equations into some sort of system form and then use the sympy.lambdify function to return a new function that I can pass to odeint
So can anyone fill in the blanks here on how I set up this ode1,ode2 system to process with SymPy.