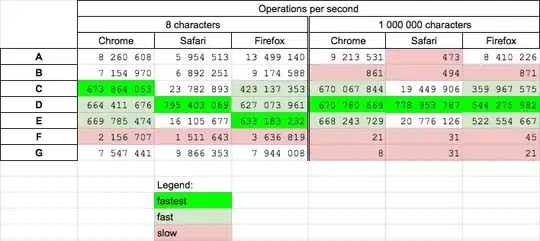
I am making several plots from a data frame using facet_wrap. My issue is with formatting the y axis. I am using pretty_breaks in scale_y_continuous to equally space the ticks on the y axis. I would now want to end the y axis on a tick as well and it doesn't seem straight forward as my plots are facetted.
So, what I have:
enter image description here
What I want:
enter image description here
Hope this makes sense! Thanks for your help
Here is the data:
ID Age Sex Genotype Organ Weight Ratio
35 1 P22 F b Body 9.2 1.00
36 1 P22 F b Heart 78.4 8.52
37 1 P22 F b Lung 156.2 16.98
38 1 P22 F b Liver 492.1 53.49
39 1 P22 F b Spleen 44.9 4.88
40 1 P22 F b Brain 313.2 34.04
41 2 P22 F a Body 9.3 1.00
42 2 P22 F a Heart 69.3 7.45
43 2 P22 F a Lung 225.6 24.26
44 2 P22 F a Liver 512.3 55.09
45 2 P22 F a Spleen 69.1 7.43
46 2 P22 F a Brain 373.2 40.13
Here is the code:
# Load file and find names
df <- read.csv('yaxis_ticks_data.csv')
# Subsetting to remove Body ratio
organ.ratios <- subset(df, df$Organ != 'Body')
## P22 Ratio plots
# Grouping for geom_point
pointGroup <- group_by(organ.ratios, Organ, Genotype, ID, Ratio)
pointGroup.Summary <- summarise(pointGroup,
n = n())
pointGroup.Summary
# Grouping for geom_bar
barGroup <- group_by(pointGroup.Summary, Organ, Genotype)
barGroup
barGroup.Summary <- summarise(barGroup,
mean_Ratio = mean(Ratio),
n = n())
barGroup.Summary
# Plot
P22.Organ.Body.plot <- ggplot() +
geom_bar(data = barGroup.Summary,
aes(x = barGroup.Summary$Genotype,
y = barGroup.Summary$mean_Ratio,
colour = Genotype,
fill = Genotype),
position = position_dodge(width = 0.9),
stat = 'identity',
show.legend = T) +
geom_point(data = pointGroup.Summary,
aes(x = pointGroup.Summary$Genotype,
y = pointGroup.Summary$Ratio,
colour = Genotype),
position = position_jitterdodge(jitter.width = 0.2, dodge.width = 0.9),
stat = 'identity',
show.legend = FALSE) +
facet_wrap(~Organ, scales = 'free', nrow = 1) +
xlab(expression(bold('Genotype'))) +
ylab(expression(bold('Organ/Body Ratio'))) +
theme(axis.line = element_line(colour = 'black'),
strip.background = element_blank(),
strip.text = element_text(face = 'bold'),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
legend.text.align = 0,
legend.title = element_text(face = 'bold'),
text = element_text(family = 'Arial', size = 14)) +
scale_color_manual('Genotype',
labels = c('a', 'b'),
values = c('black', 'black')) +
scale_fill_manual('Genotype',
labels = c('a', 'b'),
values = c('white', 'deepskyblue3')) +
scale_y_continuous(breaks = pretty_breaks(), expand = expand_scale(mult = c(0, 0.1)))
#Get plot
P22.Organ.Body.plot