I have these two codes:
v <- melt(tvalB)
data1 <- as.data.frame(cbind(tval, v$value))
sizez <- 14
ggplot(data = data1, aes(x =T-value)) +
geom_density(aes(tval, fill = "Actual"), alpha = 0.5, adjust=8) +
geom_density(aes(V2, fill = "Simulation"), alpha = 0.5, adjust=8) +
scale_colour_manual("", breaks = c("Actual", "Simulation"),
values = c("red", "blue")) +
scale_y_continuous("density") +
scale_x_continuous("t(alpha)", limits = c(-6.5,6.5)) +
labs(title ="") +
theme(plot.title = element_text(hjust = 0.5)) +
guides(fill=guide_legend(title=NULL)) +
theme(legend.title=element_text(size=sizez+1),legend.text=element_text(size=sizez),
axis.text=element_text(size=sizez-1), axis.title=element_text(size=sizez),
legend.key = element_rect(size = 0.05),
legend.key.height = unit(1.25, "cm"),
legend.key.width = unit(1.25, "cm"))
and
data1 %>%
gather(key, value) %>%
ggplot(aes(value, color=key)) +
stat_ecdf(size=0.8)+ xlim(-4.5,4.5) +
labs(x = "t(alpha)", y = "probability", color=NULL) +
scale_colour_discrete(labels = c("Actual","Simulation")) +
theme(legend.title=element_text(size=sizez+1), legend.text=element_text(size=sizez),
axis.text=element_text(size=sizez-1), axis.title=element_text(size=sizez),
legend.key = element_rect(size = 4),
legend.key.height = unit(1.3, "cm"),
legend.key.width = unit(1.3, "cm"))data1 %>%
gather(key, value) %>%
ggplot(aes(value, color=key)) +
stat_ecdf(size=0.8)+ xlim(-4.5,4.5) +
labs(x = "t(alpha)", y = "probability", color=NULL) +
scale_colour_discrete(labels = c("Actual","Simulation")) +
theme(legend.title=element_text(size=sizez+1), legend.text=element_text(size=sizez),
axis.text=element_text(size=sizez-1), axis.title=element_text(size=sizez),
legend.key = element_rect(size = 4),
legend.key.height = unit(1.3, "cm"),
legend.key.width = unit(1.3, "cm"))
It produces these two plots:
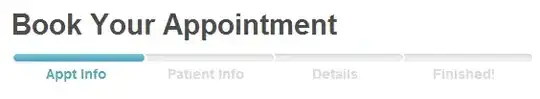
As it appears from the pictures, the gray area in the two plots doesn`t have equal heights. Is there some way to predefine the plot margins, such that if the plots are placed next to each other, in a report, they are identical in height, etc. Thank you in advance!!