EDIT: Problem solved, keeping this open for posterity.
numpy.genfrontext has trouble delimiting strings that have commas. In order to solve this, simply use pandas.read_csv and employ the quotechar = '"' to allow the importer to properly handle the strings that contain your commas.
Strange problem here.
I'm importing lists of protein data from .csv files that, for 99.9% of IDs, works flawlessly. However, 1 ID out of ~5000 thousand IDs is consistently importing the wrong data.
Here's the code I use to pull in my data. It's using glob to pull in csv files with similar names. Headers are stored as a list and then used as columns just in case the csv files have their headers mixed around (damn you, Proteome Discoverer):
indexes = ["Accession", "# Peptides", "MW [kDa]", "Score"]
headers = pd.read_csv(str(WorkingDirectory) + "/" + str(name) + "-R1.csv", nrows=1).columns.tolist()
total = [np.genfromtxt(x, delimiter = ',', skip_header = 1, usecols = [int(headers.index(indexes[0])),int(headers.index(indexes[1])),int(headers.index(indexes[2])),int(headers.index(indexes[3]))], filling_values = 0.01, dtype = ('|U16','float64','float64','float64')).tolist() for x in glob.glob(str(WorkingDirectory) + "/" + str(name) + "*.csv")]
IDs are then stored in a list, where each list entry matches the original file. [File 1, File 2, File 3]
Here's where it gets weird. Of the 5.5K entries in each .csv file, there is one ID that consistently (with code restarts) reports the wrong numbers.
Please find attached the output of my program, alongside the excel sheets that the data are sourced from. Columns A, C, E and H are my imports (Accession, Score, # Peptides and MW [kDa] respectively, in orange) 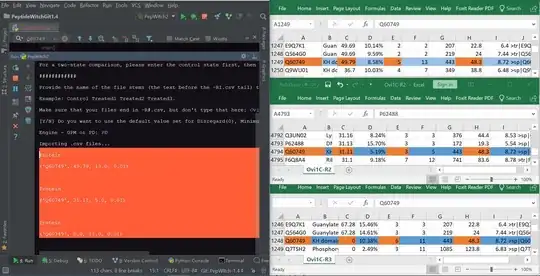
It looks like the ID's name and score are importing the correct value, but the next two columns are, respectively, off by 1 (it's importing F, not E) and then trying to pick up a value from an unspecified column that doesn't exist (hence the 0.01 due to filling values)
Things I have checked:
1) Yes, the excel headers are the same for all three files.
2) Yes, I have code in place to handle the downstream NaN nonsense that any zeros generate. So if it imports a 0 for the score, I manually change that later.
3) Yes, if there are missing values, the genfromtext filling_values = 0.01 is going to fill that gap, however for this case, it shouldn't need to fill in any gaps as there are corresponding values within the cells.
4) Every other ID I have checked is importing the data correctly.
5) Q60749 is not an unusual string. Others include: Q9CQM5, D3Z5X0, etc. No hashtags, no quote marks, no commas.
6) {From comments} All files contain only a single instance of this Protein ID
Why is this one ID causing problems out of the thousands of other successful hits? I originally found this hit because some downstream analysis said I had a NaN value; Q60749 turned out to be that value, and it's simply not importing the correct data.