Well, community:
Recently I have asked how to do exponential regression (Exponential regression function Python) thinking that for that data set the optimal regression was the Hyperbolic.
x_data = np.arange(0, 51)
y_data = np.array([0.001, 0.199, 0.394, 0.556, 0.797, 0.891, 1.171, 1.128, 1.437,
1.525, 1.720, 1.703, 1.895, 2.003, 2.108, 2.408, 2.424,2.537,
2.647, 2.740, 2.957, 2.58, 3.156, 3.051, 3.043, 3.353, 3.400,
3.606, 3.659, 3.671, 3.750, 3.827, 3.902, 3.976, 4.048, 4.018,
4.286, 4.353, 4.418, 4.382, 4.444, 4.485, 4.465, 4.600, 4.681,
4.737, 4.792, 4.845, 4.909, 4.919, 5.100])
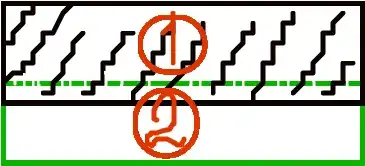
Now, I'm doubting:
The first is an exponential fit. The second is hyperbolic. I don't know which is better... How to determine? Which criteria should I follow? Is there some python function?
Thanks in advance!