I have text file (x_train) from sensor data such as accelerometers:
# (patient number, time in ms, normalization of X Y and Z,
# kurtosis, skewness, pitch, roll and yaw, label) respectively.
1,15,-0.248010047716,0.00378335508419,-0.0152548459993,-86.3738760481,0.872322164158,-3.51314800063,0
1,31,-0.248010047716,0.00378335508419,-0.0152548459993,-86.3738760481,0.872322164158,-3.51314800063,0
1,46,-0.267422664673,0.0051143782875,-0.0191247001961,-85.7662354031,1.0928406847,-4.08015176908,0
1,62,-0.267422664673,0.0051143782875,-0.0191247001961,-85.7662354031,1.0928406847,-4.08015176908,0
And i am working on a deep learning model RNN-LSTM with keras I am trying to detect if a patient is in a FOG (freezing of gait) stage or not
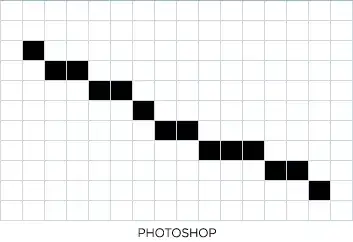
In the figure below is the chunks that i want to determine from the accelerometer signal file.
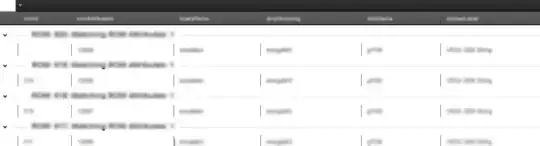
and this is the x-axis, y-axis and z-axis
The issue I am having now is that I can't figure out how to get those chunks programatically.
And also what I basically want is to know how often a patient have FOG or walking during a certain time window. (Window size around 3 seconds).
this is what i have tried
def rwindows(a, window):
shape = a.shape[0] - window + 1, window, a.shape[-1]
strides = (a.strides[0],) + a.strides
windows = np.lib.stride_tricks.as_strided(a, shape=shape, strides=strides)
return np.squeeze(windows)
s=x_train.reshape(-1,6)
print(rwindows(s,3))
and i need to obtain the difference between signals in the case of fog and walking