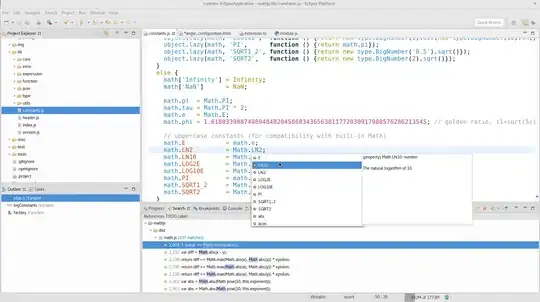
I am unable to get the labels to fit properly over each dodged bar in this chart:
I feel that I am almost there, but can't quite figure out how to get the labels perfectly positioned over each respective dodged bar.
Code:
ggplot() +
geom_col(data = leads_over_chats, aes(x = date, y = count, fill = type),
colour = "black",
position = "dodge") +
labs(title = "Leads Over Chats\n(May 2018)",
x = "Type",
y = "Count") +
geom_text(data = leads_over_chats, aes(x = date, y = count, label = count),
hjust = -1.5,
vjust = -0.5,
size = 4,
angle = 90,
position=position_dodge(width = 2.25),
colour = "black")
I am trying to replicate this (from Kibana):
Reproducible data frame
structure(list(type = structure(c(3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L, 4L,
4L, 4L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 4L, 4L, 4L, 4L,
4L, 4L, 4L, 4L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 1L, 1L, 4L, 4L, 4L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 4L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 1L, 2L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L), .Label = c("aborted-live-lead",
"conversation-claimed", "conversation-created", "lead-created"
), class = "factor"), date = structure(c(1525129200, 1525215600,
1525302000, 1525388400, 1525474800, 1525561200, 1525647600, 1525734000,
1525820400, 1525906800, 1525993200, 1526079600, 1526166000, 1526252400,
1526338800, 1526425200, 1526511600, 1526598000, 1526684400, 1526770800,
1526857200, 1526943600, 1527030000, 1527116400, 1527202800, 1527289200,
1527375600, 1527462000, 1527548400, 1527634800, 1527721200, 1526166000,
1526252400, 1526338800, 1526425200, 1526598000, 1526684400, 1526770800,
1526857200, 1526943600, 1527030000, 1527116400, 1527202800, 1527289200,
1527375600, 1527462000, 1527548400, 1527634800, 1527721200, 1526252400,
1526338800, 1526425200, 1526511600, 1526598000, 1526684400, 1526770800,
1526857200, 1526943600, 1527030000, 1527116400, 1527202800, 1527289200,
1527375600, 1527462000, 1527548400, 1527634800, 1527721200, 1526252400,
1526338800, 1526425200, 1526511600, 1526598000, 1526684400, 1526770800,
1526857200, 1526943600, 1527030000, 1527116400, 1527202800, 1527289200,
1527375600, 1527462000, 1527548400, 1527634800, 1527721200, 1525129200,
1525215600, 1525302000, 1525388400, 1525474800, 1525561200, 1525647600,
1525734000, 1525820400, 1525906800, 1525993200, 1526079600, 1526166000,
1526252400, 1526338800, 1526425200, 1526511600, 1526598000, 1526684400,
1526770800, 1526857200, 1526943600, 1527030000, 1527116400, 1527202800,
1527289200, 1527375600, 1527462000, 1527548400, 1527634800, 1527721200,
1526166000, 1526252400, 1526338800, 1526425200, 1526511600, 1526598000,
1526684400, 1526770800, 1526857200, 1526943600, 1527030000, 1527116400,
1527202800, 1527289200, 1527375600, 1527462000, 1527548400, 1527634800,
1527721200, 1526252400, 1526338800, 1526425200, 1526511600, 1526598000,
1526684400, 1526770800, 1526857200, 1526943600, 1527030000, 1527116400,
1527202800, 1527289200, 1527375600, 1527462000, 1527548400, 1527634800,
1527721200, 1526252400, 1526338800, 1526425200, 1526511600, 1526598000,
1526684400, 1526770800, 1526857200, 1526943600, 1527030000, 1527116400,
1527202800, 1527289200, 1527375600, 1527462000, 1527548400, 1527634800,
1527721200, 1525129200, 1525215600, 1525302000, 1525388400, 1525474800,
1525561200, 1525647600, 1525734000, 1525820400, 1525906800, 1525993200,
1526079600, 1526166000, 1526252400, 1526338800, 1526425200, 1526511600,
1526598000, 1526684400, 1526770800, 1526857200, 1526943600, 1527030000,
1527116400, 1527202800, 1527289200, 1527375600, 1527462000, 1527548400,
1527634800, 1527721200, 1525129200, 1525215600, 1525302000, 1525388400,
1525474800, 1525561200, 1525647600, 1525820400, 1525906800, 1526079600,
1526166000, 1526252400, 1526338800, 1526425200, 1526511600, 1526598000,
1526684400, 1526857200, 1526943600, 1527030000, 1527116400, 1527289200,
1527462000, 1527548400, 1527634800, 1526252400, 1526338800, 1526425200,
1526511600, 1526598000, 1526684400, 1526857200, 1526943600, 1527030000,
1527116400, 1527202800, 1527289200, 1527462000, 1527548400, 1527634800,
1525129200, 1525215600, 1525302000, 1525388400, 1525734000, 1525820400,
1525906800, 1525993200, 1526252400, 1526425200, 1526511600, 1526598000,
1526684400, 1526857200, 1527030000, 1527116400, 1527202800, 1527289200,
1527548400, 1527721200, 1525129200, 1525215600, 1525302000, 1525388400,
1525474800, 1525647600, 1525820400, 1525906800, 1525993200, 1526079600,
1526252400, 1526857200, 1526943600, 1527030000, 1527116400, 1527202800,
1527462000, 1527548400, 1527634800, 1527721200, 1526857200, 1526943600,
1527030000, 1527116400, 1527202800, 1527462000, 1527548400, 1527634800,
1527721200, 1526252400, 1526684400, 1526857200, 1526943600, 1527030000,
1527202800, 1527462000, 1527548400, 1526252400, 1526857200, 1526943600,
1527030000, 1527548400, 1525215600, 1525302000, 1525388400, 1525474800,
1525647600, 1525820400, 1525993200, 1526079600, 1526252400, 1526338800,
1526511600, 1526598000, 1526684400, 1526770800, 1526857200, 1526943600,
1527030000, 1527116400, 1527289200, 1527462000, 1527634800, 1527721200,
1526425200, 1526857200, 1526943600, 1527202800, 1526425200, 1527202800,
1526425200, 1526943600, 1527202800, 1526857200, 1527202800, 1526338800,
1526425200, 1526943600, 1527116400, 1527289200, 1527721200, 1525734000,
1526338800, 1526425200, 1526943600, 1527116400, 1527202800, 1527289200,
1527202800, 1525302000, 1525388400, 1525734000, 1525820400, 1525993200,
1526511600, 1526857200, 1526943600, 1527116400, 1527462000, 1527548400,
1525129200, 1525215600, 1525561200, 1525734000, 1525906800, 1526079600,
1526252400, 1526857200, 1526943600, 1527375600, 1525302000, 1525474800,
1525993200, 1526425200, 1527030000, 1525215600, 1525734000, 1526425200,
1526857200, 1527548400, 1525734000, 1526943600, 1525906800, 1526943600,
1526252400, 1526338800, 1525215600, 1525820400, 1526252400, 1527202800,
1525215600, 1526338800, 1526511600, 1526857200, 1525129200, 1527116400
), class = c("POSIXct", "POSIXt"), tzone = ""), count = c(76L,
82L, 64L, 59L, 70L, 30L, 42L, 54L, 61L, 77L, 83L, 79L, 92L, 120L,
99L, 145L, 2L, 70L, 88L, 79L, 119L, 101L, 133L, 147L, 177L, 96L,
93L, 108L, 137L, 112L, 107L, 15L, 89L, 68L, 85L, 34L, 45L, 44L,
64L, 54L, 54L, 62L, 114L, 52L, 31L, 54L, 56L, 57L, 45L, 27L,
20L, 33L, 1L, 25L, 29L, 21L, 29L, 26L, 41L, 54L, 51L, 32L, 33L,
33L, 51L, 33L, 36L, 23L, 28L, 46L, 2L, 24L, 24L, 18L, 38L, 36L,
47L, 44L, 39L, 24L, 37L, 27L, 50L, 29L, 39L, 6L, 10L, 2L, 11L,
1L, 5L, 9L, 8L, 21L, 17L, 18L, 24L, 8L, 20L, 19L, 22L, 22L, 20L,
21L, 16L, 26L, 23L, 23L, 19L, 36L, 17L, 14L, 31L, 33L, 28L, 28L,
3L, 18L, 7L, 9L, 12L, 13L, 1L, 4L, 13L, 8L, 8L, 6L, 18L, 8L,
4L, 3L, 15L, 13L, 16L, 5L, 11L, 9L, 5L, 10L, 9L, 6L, 7L, 10L,
14L, 8L, 11L, 9L, 9L, 18L, 9L, 8L, 6L, 2L, 5L, 6L, 7L, 3L, 11L,
5L, 11L, 6L, 5L, 10L, 8L, 3L, 3L, 12L, 14L, 11L, 13L, 12L, 19L,
24L, 15L, 17L, 15L, 19L, 19L, 18L, 10L, 15L, 23L, 30L, 20L, 21L,
10L, 21L, 24L, 17L, 14L, 14L, 12L, 14L, 22L, 22L, 32L, 38L, 9L,
23L, 12L, 16L, 1L, 1L, 2L, 5L, 1L, 1L, 2L, 4L, 3L, 5L, 3L, 1L,
3L, 1L, 2L, 5L, 2L, 5L, 2L, 1L, 2L, 2L, 1L, 3L, 1L, 1L, 6L, 2L,
4L, 7L, 4L, 6L, 2L, 1L, 3L, 1L, 2L, 1L, 3L, 1L, 4L, 7L, 6L, 1L,
3L, 6L, 4L, 10L, 4L, 5L, 9L, 4L, 1L, 2L, 4L, 3L, 9L, 1L, 3L,
4L, 1L, 2L, 1L, 2L, 2L, 6L, 1L, 1L, 3L, 1L, 2L, 4L, 4L, 1L, 3L,
5L, 6L, 3L, 1L, 1L, 2L, 1L, 1L, 3L, 4L, 3L, 1L, 1L, 1L, 1L, 1L,
1L, 2L, 1L, 1L, 3L, 2L, 1L, 3L, 2L, 1L, 2L, 1L, 2L, 2L, 1L, 1L,
1L, 2L, 3L, 1L, 1L, 2L, 1L, 3L, 1L, 3L, 5L, 3L, 2L, 4L, 8L, 1L,
4L, 2L, 1L, 1L, 16L, 1L, 16L, 1L, 1L, 2L, 1L, 1L, 2L, 2L, 3L,
7L, 3L, 1L, 1L, 1L, 1L, 3L, 3L, 1L, 1L, 1L, 2L, 1L, 1L, 2L, 3L,
1L, 5L, 6L, 1L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L,
1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 1L,
2L, 2L, 1L, 1L, 2L, 1L, 1L, 1L, 1L, 1L)), class = "data.frame", row.names = c(NA,
-398L))