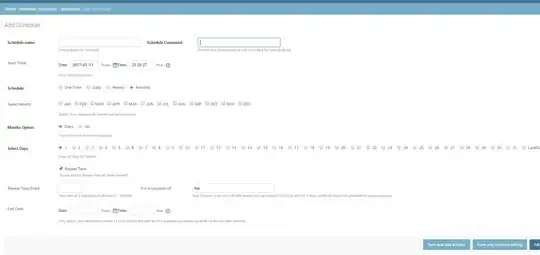
I'm trying to replicate a plot, created in gnuplot, using matplotlib. The type of plot used by gnuplot (below) is called a pseudo 3D plot.
What this is showing is secondary structure assignment for each residue of a peptide/protein over time. So, at each time step (column) you're seeing the secondary structure assignment for a single peptide. For each residue (row) you're seeing how that residue's secondary structure assignment changes over time.
I want to adapt this type of plot to matplotlib. What would be an appropriate type of plot in matplotlib to do this?
The most rudimentry solution I could think of was to simply use patches to draw rectangles at the correct coordinates, but I was hoping to use an existing matplotlib plot type that's more flexible and robust to customisation.
The data is structured like so:
data = [[x1, y1, z11], [x1, y2, z12], [x1, y3, z13], ..., [xn, ym, znm]]
Where n is the total number of time steps and m is the total number of residues. Z values are categorical data for secondary structure assignment at the xy coordinates.
Some actual data you could use to test this where there is a peptide with three residues over three time steps:
data = [[1, 1, 0], [1, 2, 0], [1, 3, 0], [2, 1, 6], [2, 2, 6], [2, 3, 6], [3, 1, 6], [3, 2, 0], [3, 3, 0]]