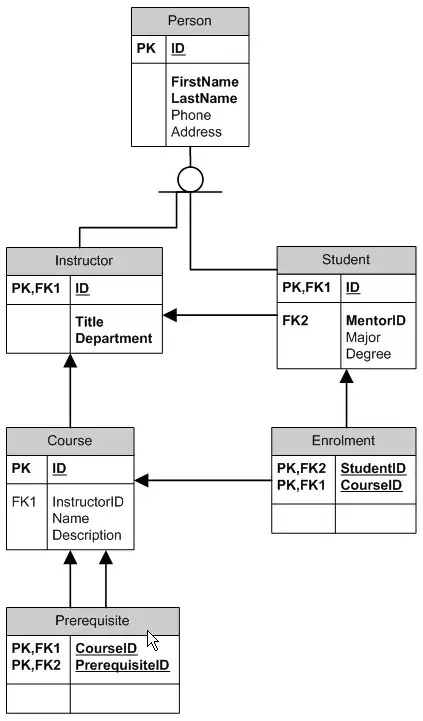
I am facing an issue. I want to plot all four variables in RStudio. Where I appear to have 2 groups for 3 variables and a Count. Yet do not have a clue how to do this with ggplot2. On xlim axes shall be age_band and sex. On y axis Count of those admitted and not admitted. I want the legend bellow the overlayed barplot. Bellow I have added the drawn picture due to confidentiality of the analysis and data. Can someone help? I've searched on stackoverflow and could not find a good reproducible code
And here is 2 types of data I have after manipulation techniques.
First type of data:
structure(list(age_band = c("0 yrs", "0 yrs", "0 yrs", "0 yrs",
"1-4 yrs", "1-4 yrs", "1-4 yrs", "1-4 yrs",
"10-14 yrs", "10-14 yrs", "10-14 yrs", "10-14 yrs",
"15-19 yrs", "15-19 yrs", "15-19 yrs","15-19 yrs"),
sex = c("Female", "Female", "Male", "Male", "Female",
"Female", "Male", "Male", "Female", "Female",
"Male", "Male", "Female", "Female", "Male", "Male"),
patient.class = c("Not Admitted", "ORDINARY ADMISSION",
"Not Admitted", "ORDINARY ADMISSION", "Not
Admitted", "ORDINARY ADMISSION", "Not
Admitted", "ORDINARY ADMISSION",
"Not Admitted", "ORDINARY ADMISSION", "Not
Admitted", "ORDINARY ADMISSION", "Not
Admitted", "ORDINARY ADMISSION",
"Not Admitted", "ORDINARY ADMISSION"),
Count = c(5681L, 1458L, 7667L, 2154L, 8040L, 2481L, 11737L,
3601L, 2904L, 938L, 3883L, 1233L, 3251L, 1266L,
2465L, 1031L)),
row.names = c(NA, -16L), class = c("tbl_df", "tbl",
"data.frame"
))
Second type of data:
structure(list(age_band = c("0 yrs", "0 yrs", "0 yrs", "0 yrs",
"1-4 yrs", "1-4 yrs", "1-4 yrs", "1-4 yrs",
"10-14 yrs", "10-14 yrs",
"10-14 yrs", "10-14 yrs", "15-19 yrs",
"15- 19 yrs", "15-19 yrs", "15-19 yrs"),
sex_patient_class = c("female_admitted", "female_not_admitted",
"male_admitted", "male_not_admitted",
"female_admitted", "female_not_admitted",
"male_admitted", "male_not_admitted",
"female_admitted", "female_not_admitted",
"male_admitted", "male_not_admitted",
"female_admitted", "female_not_admitted",
"male_admitted", "male_not_admitted"),
Count = c(1458L, 5681L, 2154L, 7667L, 2481L, 8040L, 3601L, 11737L,
938L, 2904L, 1233L, 3883L, 1266L, 3251L, 1031L, 2465L)),
row.names = c(NA, -16L), class = c("grouped_df", "tbl_df", "tbl",
"data.frame"),
vars = "age_band", drop = TRUE, indices = list( 0:3, 4:7, 8:11,
12:15),
group_sizes = c(4L, 4L, 4L, 4L), biggest_group_size = 4L, labels =
structure(list(age_band = c("0 yrs", "1-4 yrs", "10-14 yrs", "15-19
yrs")),
row.names = c(NA, -4L), class = "data.frame", vars = "age_band",
drop = TRUE))