I have a dataframe as below (obtained after lot of preprocessing)
Please find dataframe
d = {'token': {361: '180816_031', 119: '180816_031', 101: '180816_031', 135: '180816_031', 292: '180816_031',
133: '180816_031', 99: '180816_031', 270: '180816_031', 19: '180816_031', 382: '180816_031',
414: '180816_031', 267: '180816_031', 218: '180816_031', 398: '180816_031', 287: '180816_031',
155: '180816_031', 392: '180816_031', 265: '180816_031', 239: '180816_031', 237: '180816_031'},
'station': {361: 'deneb', 119: 'callisto', 101: 'callisto', 135: 'callisto', 292: 'callisto', 133: 'deneb',
99: 'callisto', 270: 'callisto', 19: 'deneb', 382: 'callisto', 414: 'deneb', 267: 'callisto',
218: 'deneb', 398: 'callisto', 287: 'deneb', 155: 'deneb', 392: 'deneb', 265: 'callisto',
239: 'callisto', 237: 'callisto'},
'cycle_number': {361: 'cycle09', 119: 'cycle06', 101: 'cycle04', 135: 'cycle01', 292: 'cycle04', 133: 'cycle05',
99: 'cycle06', 270: 'cycle07', 19: 'cycle04', 382: 'cycle08', 414: 'cycle04', 267: 'cycle10',
218: 'cycle07', 398: 'cycle08', 287: 'cycle09', 155: 'cycle08', 392: 'cycle06', 265: 'cycle02',
239: 'cycle09', 237: 'cycle07'},
'variable': {361: 'adj_high_quality_reads', 119: 'short_pass', 101: 'short_pass', 135: 'cell_mask_bilayers_sum',
292: 'adj_active_polymerase', 133: 'cell_mask_bilayers_sum', 99: 'short_pass',
270: 'adj_active_polymerase', 19: 'Unnamed: 0', 382: 'adj_high_quality_reads',
414: 'num_align_high_quality_reads', 267: 'adj_active_polymerase', 218: 'adj_single_pores',
398: 'num_align_high_quality_reads', 287: 'adj_active_polymerase', 155: 'cell_mask_bilayers_sum',
392: 'num_align_high_quality_reads', 265: 'adj_active_polymerase', 239: 'adj_single_pores',
237: 'adj_single_pores'},
'value': {361: 99704.0, 119: 2072785.0, 101: 2061059.0, 135: 1682208.0, 292: 675306.0, 133: 1714292.0,
99: 2072785.0, 270: 687988.0, 19: 19.0, 382: np.nan, 414: 285176.0, 267: 86914.0, 218: 948971.0,
398: 405196.0, 287: 137926.0, 155: 1830032.0, 392: 480081.0, 265: 951689.0, 239: 681452.0,
237: 882671.0}}
Data:
token station cycle_number variable \
19 180816_031 deneb cycle04 Unnamed: 0
99 180816_031 callisto cycle06 short_pass
101 180816_031 callisto cycle04 short_pass
119 180816_031 callisto cycle06 short_pass
133 180816_031 deneb cycle05 cell_mask_bilayers_sum
135 180816_031 callisto cycle01 cell_mask_bilayers_sum
155 180816_031 deneb cycle08 cell_mask_bilayers_sum
218 180816_031 deneb cycle07 adj_single_pores
237 180816_031 callisto cycle07 adj_single_pores
239 180816_031 callisto cycle09 adj_single_pores
265 180816_031 callisto cycle02 adj_active_polymerase
267 180816_031 callisto cycle10 adj_active_polymerase
270 180816_031 callisto cycle07 adj_active_polymerase
287 180816_031 deneb cycle09 adj_active_polymerase
292 180816_031 callisto cycle04 adj_active_polymerase
361 180816_031 deneb cycle09 adj_high_quality_reads
382 180816_031 callisto cycle08 adj_high_quality_reads
392 180816_031 deneb cycle06 num_align_high_quality_reads
398 180816_031 callisto cycle08 num_align_high_quality_reads
414 180816_031 deneb cycle04 num_align_high_quality_reads
value
19 19.0
99 2072785.0
101 2061059.0
119 2072785.0
133 1714292.0
135 1682208.0
155 1830032.0
218 948971.0
237 882671.0
239 681452.0
265 951689.0
267 86914.0
270 687988.0
287 137926.0
292 675306.0
361 99704.0
382 NaN
392 480081.0
398 405196.0
414 285176.0
I am trying to create scatterplot with smooth line (Expected Output below)
I am using below code (with help) to replicate the same, however I am having my legend values overlapped in the plotting area.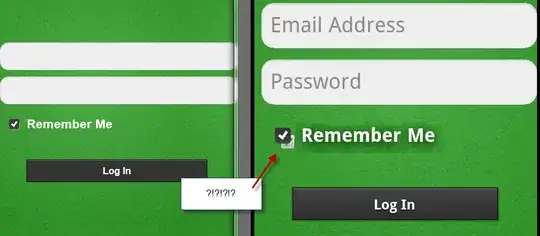
Code Used to produce Output
df['cycle_number'] = df['cycle_number'].str.replace('cycle', '')
df['cycle_number'] = df['cycle_number'].apply(pd.to_numeric)
fig, ax = plt.subplots()
fig.set_size_inches(16, 4)
# sns.pointplot('cycle_number', 'value', data=df, hue='variable', err_style="bars", ci=68)
g2=sns.lmplot('cycle_number', 'value', data=df, hue='variable', ci=2, order=5, truncate=True)
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
# Put a legend to the right of the current axis
#ax.legend(loc='right', bbox_to_anchor=(1, 0.1))
ax.legend(loc='left', bbox_to_anchor=(1, 0))
for p in ax.patches:
ax.annotate("%.2f" % p.get_height(), (p.get_x() + p.get_width() / 2., p.get_height()),
ha='center', va='center', fontsize=11, color='gray', xytext=(0, 20),
textcoords='offset points')
plt.show()
Please help to remove the overlapped legend from plotting area