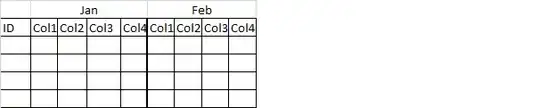
I am trying to create some correlation plots based of a data frame that I created using dplyr's spread() function. When I used the spread function, it created NAs in the new data frame. This makes sense because the data frame had concentration values for different parameters at different time periods.
Here is an example screenshot of the original data frame:
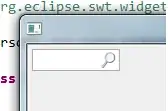
When I used the spread function it gave me a data frame like this(sample data):
structure(list(orgid = c("11NPSWRD", "11NPSWRD", "11NPSWRD",
"11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD",
"11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD",
"11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD", "11NPSWRD"),
locid = c("11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2",
"11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2",
"11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2",
"11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2",
"11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2",
"11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2",
"11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2", "11NPSWRD-MORR_NPS_PR2"
), stdate = structure(c(9891, 9891, 9891, 9920, 9920, 9920,
9949, 9949, 9949, 9978, 9978, 9978, 10011, 10011, 10011,
10067, 10067, 10073, 10073, 10073), class = "Date"), sttime = structure(c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), class = c("hms",
"difftime"), units = "secs"), valunit = c("uS/cm", "mg/l",
"mg/l", "uS/cm", "mg/l", "mg/l", "uS/cm", "mg/l", "mg/l",
"uS/cm", "mg/l", "mg/l", "uS/cm", "mg/l", "mg/l", "uS/cm",
"mg/l", "uS/cm", "mg/l", "mg/l"), swqs = c("FW2-TP", "FW2-TP",
"FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP",
"FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP",
"FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP", "FW2-TP"
), WMA = c(6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L,
6L, 6L, 6L, 6L, 6L, 6L, 6L, 6L), year = c(1997L, 1997L, 1997L,
1997L, 1997L, 1997L, 1997L, 1997L, 1997L, 1997L, 1997L, 1997L,
1997L, 1997L, 1997L, 1997L, 1997L, 1997L, 1997L, 1997L),
Chloride = c(NA, 35, NA, NA, 45, NA, NA, 30, NA, NA, 30,
NA, NA, 30, NA, NA, NA, NA, 35, NA), `Specific conductance` = c(224,
NA, NA, 248, NA, NA, 204, NA, NA, 166, NA, NA, 189, NA, NA,
119, NA, 194, NA, NA), `Total dissolved solids` = c(NA, NA,
101, NA, NA, 115, NA, NA, 96, NA, NA, 79, NA, NA, 89, NA,
56, NA, NA, 92)), .Names = c("orgid", "locid", "stdate",
"sttime", "valunit", "swqs", "WMA", "year", "Chloride", "Specific conductance",
"Total dissolved solids"), row.names = c(NA, 20L), class = "data.frame")
The problem I am having is when I try and create the correlation plot it's giving me a plot with only one point.. I'm guessing this is because there are NAs in the data frame.. But when I try and filter the NAs it gives me a data frame with 0 observations.. Any help would be greatly appreciated!!
Example code to create correlation plot:
plot1<-ggplot(data=df,aes(x="Specific conductance",y="Chloride"))+
geom_smooth(method = "lm", se=FALSE, color="black", formula = y ~ x)+
geom_point()