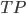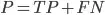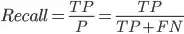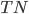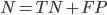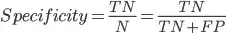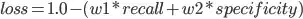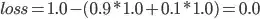I'll briefly introduce the concepts we're trying to tackle.
Recall
From all that were positive, how many did our model predict as positive?
All that were positive = 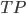
What our model said were positive = 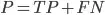
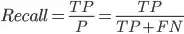
Since recall is inversely proportional to FN, improving it decreases FN.
Specificity
From all that were negative, how many did our model predict as negative?
All that were negative = 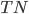
What our model said were negative = 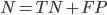
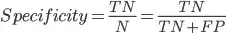
Since specificity is inversely proportional to FP, improving it decreases FP.
In your next searches, or whatever classification-related activity you perform, knowing these is going to give you an extra edge in communication and understanding.
A Solution
So. These two concepts, as you mas have figured out already, are opposites. This means that increasing one is likely to decrease the other.
Since you want priority on recall, but don't want to loose too much in specificity, you can combine both of those and attribute weights. Following what's clearly explained in this answer:
import numpy as np
import keras.backend as K
def binary_recall_specificity(y_true, y_pred, recall_weight, spec_weight):
TN = np.logical_and(K.eval(y_true) == 0, K.eval(y_pred) == 0)
TP = np.logical_and(K.eval(y_true) == 1, K.eval(y_pred) == 1)
FP = np.logical_and(K.eval(y_true) == 0, K.eval(y_pred) == 1)
FN = np.logical_and(K.eval(y_true) == 1, K.eval(y_pred) == 0)
# Converted as Keras Tensors
TN = K.sum(K.variable(TN))
FP = K.sum(K.variable(FP))
specificity = TN / (TN + FP + K.epsilon())
recall = TP / (TP + FN + K.epsilon())
return 1.0 - (recall_weight*recall + spec_weight*specificity)
Notice recall_weight and spec_weight? They're weights we're attributing to each of the metrics. For distribution convention, they should always add to 1.0¹, e.g. recall_weight=0.9, specificity_weight=0.1. The intention here is for you to see what proportion best suits your needs.
But Keras' loss functions must only receive (y_true, y_pred) as arguments, so let's define a wrapper:
# Our custom loss' wrapper
def custom_loss(recall_weight, spec_weight):
def recall_spec_loss(y_true, y_pred):
return binary_recall_specificity(y_true, y_pred, recall_weight, spec_weight)
# Returns the (y_true, y_pred) loss function
return recall_spec_loss
And onto using it, we'd have
# Build model, add layers, etc
model = my_model
# Getting our loss function for specific weights
loss = custom_loss(recall_weight=0.9, spec_weight=0.1)
# Compiling the model with such loss
model.compile(loss=loss)
¹ The weights, added, must total 1.0, because in case both recall=1.0 and specificity=1.0 (the perfect score), the formula
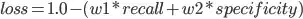
Shall give us, for example,
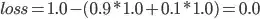
Clearly, if we got the perfect score, we'd want our loss to equal 0.