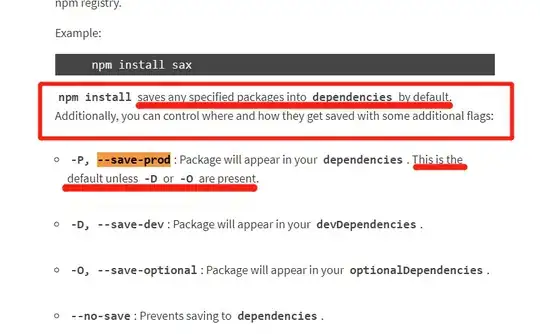
I have a data frame like so:
df<- data.frame(month= rep(c("Jan", "Feb", "Mar", "Apr", "May"), 3),
year= c(seq(2001:2003,5), rep(2002, 5), rep(2003, 5)),
clim_var= c(rep("precip_mm", 5), rep("tmin",5), rep("tmax", 5)),
anomaly= sample(-20:20, 15, replace = TRUE))
df<-df[-c(3,10),]
library("zoo")
df$date<- as.yearmon(paste(df$year, df$month), format= "%Y %b")
You will notice that some dates may be missing, but for the most part it is time series data for various climate variables. I would like each climate variable to be a facet. The y axis will plot anomaly for each level of of the clim_var column. Such that I should get a multifacted figure which each graph similar to this:
I have tried this code (modified from)
library(ggplot2)
gg<- ggplot(df, aes(x= seq_along(date), y = anomaly)) +
geom_bar(stat = 'identity', aes(fill = anomaly>0), position = 'dodge', col =
'transparent') +
theme_bw() + scale_fill_discrete(guide = 'none') +
labs(x = '', y = 'anomaly')
gg + facet_grid(clim_var~.)
gg+ scale_x_datetime(labels = date_format("%b %Y"))
The issue seems to be plotting the date. It's as though it is not being recognized as a date so data for each clim_var take up 1/3 of the plot area and the x-axis is continuous values rather than dates. I want the output to have axis label that include the month and year like so...
In my real dataset there are many years of data, so it may be most clean to specify labels for only Jan and then leave other months as tick marks without labels. Any insight to that would be appreciated.
Edit:
Corrected data frame such that each clim-var has multiple years of data
precip_mm<- data.frame(clim_var= rep("precip_mm",36), month= rep(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sept", "Oct", "Nov", "Dec" ), 3),
year= c(rep(2001,12),rep(2002,12), rep(2003, 12)),
anomaly= sample(-20:20, 36, replace = TRUE))
tmin<- data.frame(clim_var= rep("tmin",36), month= rep(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sept", "Oct", "Nov", "Dec" ), 3),
year= c(rep(2001,12),rep(2002,12), rep(2003, 12)),
anomaly= sample(-20:20, 36, replace = TRUE))
tmax<- data.frame(clim_var= rep("tmax",36), month= rep(c("Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sept", "Oct", "Nov", "Dec" ), 3),
year= c(rep(2001,12),rep(2002,12), rep(2003, 12)),
anomaly= sample(-20:20, 36, replace = TRUE))
df<- rbind(precip_mm, tmin)
df<-rbind(df, tmax)
df<-df[-c(3,10, 50, 100),]
library("zoo")
df$date<- as.yearmon(paste(df$year, df$month), format= "%Y %b")
Adjustments suggested in comments
library(ggplot2)
gg<- ggplot(df, aes(x= date, y = anomaly)) +
geom_bar(stat = 'identity', aes(fill = anomaly>0), position = 'dodge', col =
'transparent') +
theme_bw() + scale_fill_discrete(guide = 'none') +
labs(x = '', y = 'anomaly')
gg + facet_grid(clim_var~.)
gg+ scale_x_yearmon()
Output does not facet each clim_var, however x-axis is correctly labeled.
Edit2:
labels_month <- format(seq(from = min(df$date), to =
max(df$date), by = "1 months"), "%Y-%b")
labels_month[rep(c(FALSE, TRUE), c(1, 11))] <- ""
labels_month<- as.Date(labels_month, format= "%Y-%b")
x_breaks <- seq(min(df$date), max(df$date), by = "1 months")
p1 <- ggplot(df, aes(x = factor(date), y = df)) +
geom_col(aes(fill = anomoly > 0),
position = "dodge",
col = "transparent") +
theme_bw(base_size = 12) +
scale_fill_discrete(guide = "none") +
labs(x = "", y = "") +
scale_x_date(expand = c(0.015, 0.015),
labels = labels_month,
breaks = x_breaks) +
theme(axis.text.x = element_text(angle = 90, vjust = 0.5))
facet_grid(climvar ~ .,
labeller = label_parsed,
switch = "y",
scales = 'free_y')
p1