I am trying to plot 3 different plot on the same axis of scale in one plot. The plot is coming fine but y-axis scale number are overlapping each other. Here is my plot.
h1<-hazard.plot.w2p(beta=beta.spreda,eta=eta.spreda,time=exa1.dat$time,line.colour="orange")
h2<-hazard.plot.w2p(beta=1.007629,eta=32.56836,time=exa1.dat$time,line.colour="red")
h3<-hazard.plot.w2p(beta=1.104483,eta=36.53923,time=exa1.dat$time,line.colour="green")
Function used to run this code:
hazard.plot.w2p <- function(beta, eta, time, line.colour, nincr = 500) {
max.time <- max(time, na.rm = F)
t <- seq(0, max.time, length.out = nincr)
r <- numeric(length(t))
for (i in 1:length(t)) {
r[i] <- failure.rate.w2p(beta, eta, t[i])
}
plot(t, r, type = 'l', bty = 'l',
col = line.colour, lwd = 2,
main = "", xlab = "Time",
ylab = "Failure rate",
las = 1, adj = 0.5,
cex.axis = 0.85, cex.lab = 1.2)
par(new=TRUE)
}
Sample DataSet:
[fail time
a 4.55
a 4.65
a 5.21
b 3.21
a 1.21
a 5.65
a 7.12][1]
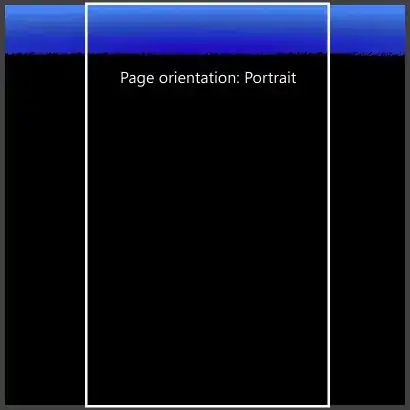
This is the output I am getting: