I'm working on a sequence forecasting problem and I don't have much experience in this area, so some of the below questions might be naive.
FYI: I've created a follow-up question with a focus on CRFs here
I have the following problem:
I would like to forecast a binary sequence for multiple, non-independent variables.
Inputs:
I have a dataset with the following variables:
- Timestamps
- Groups A and B
- Binary signal corresponding to each group at a particular timestamp
Additionally, suppose the following:
- We can extract additional attributes from the timestamps (e.g. hour of day) which can be used as external predictors
- We believe that groups A and B are not independent therefore it might be optimal to model their behaviour jointly
binary_signal_group_A and binary_signal_group_B are the 2 non-independent variables that I would like to forecast using (1) their past behaviour and (2) additional information extracted from each timestamp.
What I've done so far:
# required libraries
import re
import numpy as np
import pandas as pd
from keras import Sequential
from keras.layers import LSTM
data_length = 18 # how long our data series will be
shift_length = 3 # how long of a sequence do we want
df = (pd.DataFrame # create a sample dataframe
.from_records(np.random.randint(2, size=[data_length, 3]))
.rename(columns={0:'a', 1:'b', 2:'extra'}))
# NOTE: the 'extra' variable refers to a generic predictor such as for example 'is_weekend' indicator, it doesn't really matter what it is
# shift so that our sequences are in rows (assuming data is sorted already)
colrange = df.columns
shift_range = [_ for _ in range(-shift_length, shift_length+1) if _ != 0]
for c in colrange:
for s in shift_range:
if not (c == 'extra' and s > 0):
charge = 'next' if s > 0 else 'last' # 'next' variables is what we want to predict
formatted_s = '{0:02d}'.format(abs(s))
new_var = '{var}_{charge}_{n}'.format(var=c, charge=charge, n=formatted_s)
df[new_var] = df[c].shift(s)
# drop unnecessary variables and trim missings generated by the shift operation
df.dropna(axis=0, inplace=True)
df.drop(colrange, axis=1, inplace=True)
df = df.astype(int)
df.head() # check it out
# a_last_03 a_last_02 ... extra_last_02 extra_last_01
# 3 0 1 ... 0 1
# 4 1 0 ... 0 0
# 5 0 1 ... 1 0
# 6 0 0 ... 0 1
# 7 0 0 ... 1 0
# [5 rows x 15 columns]
# separate predictors and response
response_df_dict = {}
for g in ['a','b']:
response_df_dict[g] = df[[c for c in df.columns if 'next' in c and g in c]]
# reformat for LSTM
# the response for every row is a matrix with depth of 2 (the number of groups) and width = shift_length
# the predictors are of the same dimensions except the depth is not 2 but the number of predictors that we have
response_array_list = []
col_prefix = set([re.sub('_\d+$','',c) for c in df.columns if 'next' not in c])
for c in col_prefix:
current_array = df[[z for z in df.columns if z.startswith(c)]].values
response_array_list.append(current_array)
# reshape into samples (1), time stamps (2) and channels/variables (0)
response_array = np.array([response_df_dict['a'].values,response_df_dict['b'].values])
response_array = np.reshape(response_array, (response_array.shape[1], response_array.shape[2], response_array.shape[0]))
predictor_array = np.array(response_array_list)
predictor_array = np.reshape(predictor_array, (predictor_array.shape[1], predictor_array.shape[2], predictor_array.shape[0]))
# feed into the model
model = Sequential()
model.add(LSTM(8, input_shape=(predictor_array.shape[1],predictor_array.shape[2]), return_sequences=True)) # the number of neurons here can be anything
model.add(LSTM(2, return_sequences=True)) # should I use an activation function here? the number of neurons here must be equal to the # of groups we are predicting
model.summary()
# _________________________________________________________________
# Layer (type) Output Shape Param #
# =================================================================
# lstm_62 (LSTM) (None, 3, 8) 384
# _________________________________________________________________
# lstm_63 (LSTM) (None, 3, 2) 88
# =================================================================
# Total params: 472
# Trainable params: 472
# Non-trainable params: 0
model.compile(loss='binary_crossentropy', optimizer='adam', metrics=['accuracy']) # is it valid to use crossentropy and accuracy as metric?
model.fit(predictor_array, response_array, epochs=10, batch_size=1)
model_preds = model.predict_classes(predictor_array) # not gonna worry about train/test split here
model_preds.shape # should return (12, 3, 2) or (# of records, # of timestamps, # of groups which are a and b)
# (12, 3)
model_preds
# array([[1, 0, 0],
# [0, 0, 0],
# [1, 0, 0],
# [0, 0, 0],
# [1, 0, 0],
# [0, 0, 0],
# [0, 0, 0],
# [0, 0, 0],
# [0, 0, 0],
# [0, 0, 0],
# [1, 0, 0],
# [0, 0, 0]])
Questions:
The main question here is this: how do I get this working so that the model would forecast the next N sequences for both groups?
Additionally, I would like to ask the following questions:
- Groups A and B are expected to be cross-correlated, however, is it valid to attempt to output both A and B sequences by a single model or should I fit 2 separate models, one predicting A, the other one predicting B but both using historical A and B data as inputs?
- While my last layer in the model is an LSTM of shape (None, 3, 2), the prediction output is of shape (12, 3) when I would have expected it to be (12, 2) -- am I doing something wrong here and if so, how would I fix this?
- As far as the output LSTM layer is concerned, would it be a good idea to use an activation function here, such as sigmoid? Why/why not?
- Is it valid to use a classification type loss (binary cross-entropy) and metrics (accuracy) for optimising a sequence?
- Is an LSTM model an optimal choice here? Does anyone think that a CRF or some HMM-type model would work better here?
Many thanks!
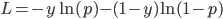
 .
If p is outside of this open interval range then the loss is undefined. The default activation of lstm layer in
.
If p is outside of this open interval range then the loss is undefined. The default activation of lstm layer in