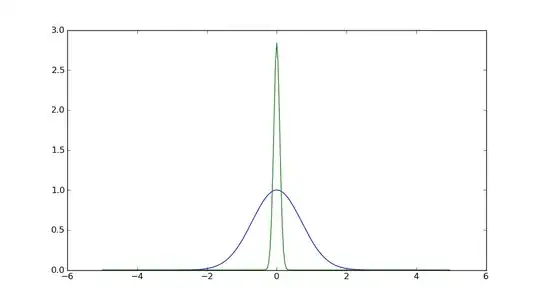
I am trying to utilize Numpy's fft function, however when I give the function a simple gausian function the fft of that gausian function is not a gausian, its close but its halved so that each half is at either end of the x axis.
The Gaussian function I'm calculating is y = exp(-x^2)
Here is my code:
from cmath import *
from numpy import multiply
from numpy.fft import fft
from pylab import plot, show
""" Basically the standard range() function but with float support """
def frange (min_value, max_value, step):
value = float(min_value)
array = []
while value < float(max_value):
array.append(value)
value += float(step)
return array
N = 256.0 # number of steps
y = []
x = frange(-5, 5, 10/N)
# fill array y with values of the Gaussian function
cache = -multiply(x, x)
for i in cache: y.append(exp(i))
Y = fft(y)
# plot the fft of the gausian function
plot(x, abs(Y))
show()
The result is not quite right, cause the FFT of a Gaussian function should be a Gaussian function itself...