I am modeling a hydrologic process (water levels [stage] in lakes measured in mm) that can be described as:
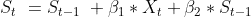
where 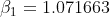 is estimated from a different model and used as a constant in this model.
is estimated from a different model and used as a constant in this model. 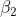 is the unknown and the value is expected to be between (-0.001,0.001). The starting value of S doesn't matter so long as it is greater than 10m (10000mm). The model runs on a daily time step. I have observed stage from multiple different lakes and fit each lake independently.
is the unknown and the value is expected to be between (-0.001,0.001). The starting value of S doesn't matter so long as it is greater than 10m (10000mm). The model runs on a daily time step. I have observed stage from multiple different lakes and fit each lake independently.
Currently, I am brute-force identifying the parameter value by:
- Creating a 100 value sequence of parameter values spanning (-0.001,0.001)
- Predicting stage using the above equation and estimate RMSE between modeled and observed data (significantly fewer observations than modeled data points)
- Identifying the B with the lowest RMSE and selecting B values on either side to create a new sequence of parameter values to search over
- Step 2 and 3 are repeated until RMSE decreases by less than 0.01 or increases.
The code for the brute force approach I've been using is below along with the data associated with a single lake.
Is there an alternative approach to estimating the unknown parameter Beta2 given the model above and the fact that I only have observed data for a limited number of days?
Thanks!
library(tidyverse)
library(lubridate)
library(Metrics)
#The Data
dat <- read_csv("https://www.dropbox.com/s/skg8wfpu9274npb/driver_data.csv?dl=1")
observeddata <- read_csv("https://www.dropbox.com/s/bhh27g5rupoqps3/observeddata.csv?dl=1") %>% select(Date,Value)
#Setup initial values and vectors
S = matrix(nrow = nrow(dat),ncol = 1) #create an empty matrix for predicted values
S[1,1] = 10*1000 #set initial value (mm)
rmse.diff <- 10^100 #random high value for difference between min RMSE between successive
#parameter searches
b.levels <- seq(from = -0.001,to = 0.001,length.out=100) #random starting parameter that should contain
#the final value being estimated
n = 0 # counter
#Loop to bruteforce search for best parameter estimate
while(rmse.diff > 0.01 ) {
rmse.vec = rep(NA,length(b.levels))
for(t in 1:length(b.levels)){
for(z in 2:nrow(S)){
S[z,1] <- S[(z-1),1] + (1.071663*(dat$X[z])) + (b.levels[t]*(S[(z-1),1])) #-1.532236
} #end of time series loop
extrap_level <- data.frame(Date= dat$Date, level = S) # predicted lake levels
#calculate an offset to center observed data on extrapolated data
dat.offset = observeddata %>% left_join(extrap_level) %>%
mutate(offset = level-Value) %>% drop_na()
offset <- mean(dat.offset$offset)
dat.compare <- observeddata %>% left_join(extrap_level) %>%
mutate(Value = Value + offset) %>% drop_na()
#calculate RMSE between observed and extrapolated values
rmse.vec[t] <- rmse(actual = dat.compare$Value,predicted = dat.compare$level)
#plot the data to watch how parameter choice influences fit while looping
#plots have a hard time keeping up
if(t ==1 | t==50 | t==100) {
plot(extrap_level$Date,extrap_level$level,type="l")
lines(dat.compare$Date,dat.compare$Value,col="red")
}
}
#find minimum RMSE value
min.rmse <- which(rmse.vec==min(rmse.vec))
if(n == 0) rmse.best <- rmse.vec[min.rmse] else rmse.best = c(rmse.best,rmse.vec[min.rmse])
if (n >= 1) rmse.diff <- (rmse.best[n]-rmse.best[n+1])
if(rmse.diff < 0) break()
best.b <- b.levels[min.rmse]
#take the parameter values on either side of the best prior RMSE and use those as search area
b.levels <- seq(from = (b.levels[min.rmse-1]),to = (b.levels[min.rmse+1]),length.out=100)
n = n + 1
}
rmse.best #vector of RMSE for each parameter search
best.b #Last identified best parameter value