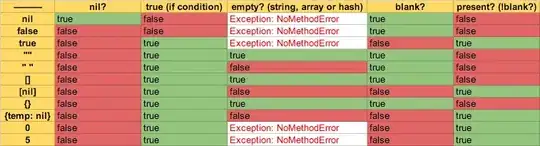
I'm trying to plot a few different histograms with the same x-axis limits. In order to do this, I've extracted the xlimits from the histogram with the widest x-axis limits and am forcing every plot to have these limits. However, when I custom set my limits to this axis range for the same data from which I extracted the limits, it removed 4 data points. They're clearly in the range of the axis limits - in fact, one is nearly right in the middle. Any idea what is causing this behavior? The code below hopefully makes clear what I'm attempting
original <- ggplot(myData, aes (x = Signal, fill = Positivity)) +
geom_histogram(alpha = 0.2, position = "identity", color = "black") +
scale_x_log10() + theme_bw() + xlab("Original Limits")
original_info<-ggplot_build(original) #get plot info
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
xlimlow <- 10^original_info$layout$panel_scales_x[[1]]$range$range[1] # get lower x axis limit, convert because log10
xlimhigh <- 10^original_info$layout$panel_scales_x[[1]]$range$range[2] # get upper x axis limit, convert because log10
hist_custom_limits <- ggplot(myData, aes (x = Signal, fill = Positivity)) +
geom_histogram(alpha = 0.2, position = "identity", color = "black") +
scale_x_log10(limits = c(xlimlow, xlimhigh)) + theme_bw() + xlab("Custom Limits")
The corresponding graphs are as follows
Here is the output of reprex to provide code that should reproduce the issue
rm(list = ls())
library(ggplot2)
myData <- data.frame(
Signal = c(258L, 290L, 470L, 167L, 133L, 183L, 2441L, 225L, 64L, 140L,
204L, 398L, 113L, 269L, 838L, 183L, 182L, 440L,
107L, 161L, 215L, 408L, 225L, 1920L, 1579L, 150L, 161L,
247L, 129L, 537L, 333L, 193L, 161L, 151L, 97L, 730L,
258L, 2234L, 129L, 226L, 86L, 343L, 107L, 183L, 226L,
236L, 1029L, 7308L, 376L, 140L, 516L, 269L, 204L,
483L, 140L, 440L, 333L),
Positivity = c(0L, 0L, 1L, 1L, 0L, 1L, 1L, 0L, 0L, 1L, 0L, 0L, 0L, 1L, 1L,
1L, 0L, 0L, 1L, 0L, 0L, 0L, 0L, 1L, 1L, 0L, 0L, 0L,
0L, 1L, 0L, 0L, 0L, 1L, 0L, 1L, 0L, 1L, 1L, 0L, 0L,
1L, 1L, 0L, 1L, 0L, 0L, 1L, 1L, 0L, 1L, 0L, 0L, 1L,
1L, 0L, 0L)
)
myData$Positivity <- factor(myData$Positivity)
levels(myData$Positivity) = c("Negative", "Positive")
original <- ggplot(myData, aes (x = Signal, fill = Positivity)) +
geom_histogram(alpha = 0.2, position = "identity", color = "black") +
scale_x_log10() + theme_bw() + xlab("Original Limits")
original_info<-ggplot_build(original) #get plot info
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
xlimlow <- 10^original_info$layout$panel_scales_x[[1]]$range$range[1] # get lower x axis limit, convert because log10
xlimhigh <- 10^original_info$layout$panel_scales_x[[1]]$range$range[2] # get upper x axis limit, convert because log10
hist_custom_limits <- ggplot(myData, aes (x = Signal, fill = Positivity)) +
geom_histogram(alpha = 0.2, position = "identity", color = "black") +
scale_x_log10(limits = c(xlimlow, xlimhigh)) + theme_bw() + xlab("Custom Limits")
Created on 2019-03-12 by the reprex package (v0.2.1)