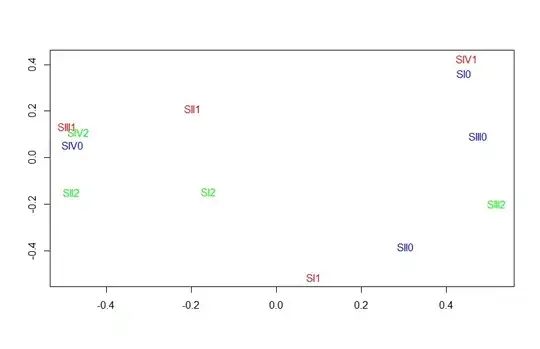
I have plotted point graph using vegan package but I want to circle the similarly treated species. As shown in the figure, 3 colors for 3 treatments. I want to circle them too.
Here is my code.
library(vegan)
library(MASS)
library(readxl)
bray1 <- read_excel("bray1.xlsx")
cols <- c("red", "blue","blue", "green","green","red","blue","green","green","red","red","blue")
row.names(bray1) <- c("SI1", "SII0", "SI0", "SII2", "SI2", "SII1", "SIII0", "SIV2", "SIII2", "SIV1", "SIII1", "SIV0")
bcdist <- vegdist(bray1, "bray")
bcmds <- isoMDS(bcdist, k = 2)
plot(bcmds$points, type = "n", xlab = "", ylab = "")
text(bcmds$points, dimnames(bray1)[[1]],col = cols,size=10)
bray1<-structure(list(`Andropogon virginicus` = c(0, 0, 0, 0, 2.7, 31.5333333333333, 0, 0, 0, 0, 0, 0), `Oenothera parviflora` = c(61.6,30.3333333333333, 7.53333333333333, 0, 11.7333333333333, 0, 0, 0,75.4, 0, 0, 0), `Lespedeza cuneata` = c(0, 0, 0, 0, 0, 46.7333333333333, 0, 0, 3, 0, 0, 0), `Lespedeza pilosa` = c(0, 1.93333333333333, 0, 0, 1.73333333333333, 0, 0, 0, 0, 1.7, 0, 0), `Chamaesyce maculata` = c(0, 0, 0,4.733333333, 0, 0, 0, 0, 0, 0, 0, 0), `Chamaesyce nutans` = c(0,0, 0, 0, 0,0, 0.166666666666667, 0, 0, 0, 0, 0), `Bidens frondosa` = c(0, 0, 0,1.76666666666667, 1.03333333333333, 3.23333333333333, 0, 0, 0, 0, 0, 0), `Erigeron annuus` = c(0, 0, 0, 0, 0.4, 0, 0, 0, 0, 0, 0, 0), `Erigeron canadensis` = c(0, 0, 0, 0, 0, 4.33333333333333, 0, 0, 9.1, 2.066666667, 0,0), `Equisetum arvense` = c(46, 62.7333333333333, 0, 1.66666666666667, 0, 0.533333333333333, 0, 0, 0, 0, 0, 0), `Erigeron sumatrensis` = c(0, 0, 0, 0, 0, 16.4333333333333, 0, 4, 0, 6.633333333, 0, 0), `Hypochaeris radicata` = c(0, 3.76666666666667, 116.6, 0, 5.033333333, 9.76666666666667, 29, 0, 23.1666666666667, 82.16666667, 0, 0), `Lactuca indica` = c(10.26666667, 0, 1.566666667, 120.1333333, 44.36666667, 42.0333333333333, 0, 14.2333333333333, 0, 0, 14.36666667, 22.2), `Solidago altissima` = c(0, 1.06666666666667, 33.93333333, 0, 0, 0, 0, 0, 0, 6.6, 0, 0), `Sonchus asper` = c(0, 35.9, 0, 0, 0, 7.46666666666667,
29.6666666666667, 4.96666666666667, 0, 0, 0.23, 2.933333333 )), .Names = c("Andropogon virginicus", "Oenothera parviflora", "Lespedeza cuneata", "Lespedeza pilosa", "Chamaesyce maculata", "Chamaesyce nutans", "Bidens frondosa", "Erigeron annuus", "Erigeron canadensis", "Equisetum arvense", "Erigeron sumatrensis", "Hypochaeris radicata", "Lactuca indica", "Solidago altissima", "Sonchus asper"), row.names = c(NA, -12L), class = c("tbl_df", "tbl", "data.frame"))