Background
I managed to reproduce the behavior:
- On Win - but this shouldn't be a problem since (I assume that) things are the same on OSX
- The Python from the (current) Anaconda environment and Anaconda's default one (which corresponds to base environment) are distinct. If this is not the case, the answer (or parts of it) might be incorrect. Note that there are the only 2 Pythons that I'm going to reference, they are both internal to Anaconda (even if the Anaconda "keyword" will not be present)
- I used PyGraphviz in my test scenarios (instead of PyOpenCL)
code0.py:
#!/usr/bin/env python3
import sys
import os
import pprint
print(f"Python Executable: {sys.executable}")
print(f"Version {sys.version} on {sys.platform}\n")
conda_env_var = "CONDA_DEFAULT_ENV"
conda_env = os.environ[conda_env_var]
print(f"{conda_env_var}: {conda_env}\n")
sys_path = pprint.pformat(sys.path)
print(f"sys.path: {sys_path}\n")
path_var = "PATH"
env_path = pprint.pformat([item for item in os.environ[path_var].split(os.pathsep) if item.find(conda_env) > -1])
print(f"os.environ[\"{path_var}\"] (relevant): {env_path}\n")
import pygraphviz
print(pygraphviz)
Output:
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> python code0.py
Python Executable: E:\Install\x64\Anaconda\Anaconda\2018.12\envs\py_064_030701_test0\python.exe
Version 3.7.1 (default, Dec 10 2018, 22:54:23) [MSC v.1915 64 bit (AMD64)] on win32
CONDA_DEFAULT_ENV: py_064_030701_test0
sys.path: ['e:\\Work\\Dev\\StackOverflow\\q055251357',
'E:\\Work\\Dev\\Utils',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\python37.zip',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\DLLs',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\lib',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\lib\\site-packages']
os.environ["PATH"] (relevant): ['E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\Library\\mingw-w64\\bin',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\Library\\usr\\bin',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\Library\\bin',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\Scripts',
'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\bin']
<module 'pygraphviz' from 'E:\\Install\\x64\\Anaconda\\Anaconda\\2018.12\\envs\\py_064_030701_test0\\lib\\site-packages\\pygraphviz\\__init__.py'>
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> where jupyter-notebook
E:\Install\x64\Anaconda\Anaconda\2018.12\Scripts\jupyter-notebook.exe
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> jupyter-notebook
[I 01:16:10.345 NotebookApp] JupyterLab extension loaded from E:\Install\x64\Anaconda\Anaconda\2018.12\lib\site-packages\jupyterlab
[I 01:16:10.346 NotebookApp] JupyterLab application directory is E:\Install\x64\Anaconda\Anaconda\2018.12\share\jupyter\lab
[I 01:16:10.349 NotebookApp] Serving notebooks from local directory: e:\Work\Dev\StackOverflow\q055251357
[I 01:16:10.350 NotebookApp] The Jupyter Notebook is running at:
[I 01:16:10.352 NotebookApp] http://localhost:8888/?token=14412a6d6d0c895d059a86bcd71e10cbface4a479c5843c2
[I 01:16:10.353 NotebookApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
[C 01:16:10.437 NotebookApp]
To access the notebook, open this file in a browser:
file:///C:/Users/cfati/AppData/Roaming/jupyter/runtime/nbserver-24700-open.html
Or copy and paste one of these URLs:
http://localhost:8888/?token=14412a6d6d0c895d059a86bcd71e10cbface4a479c5843c2
[I 01:17:18.569 NotebookApp] 302 GET /?token=14412a6d6d0c895d059a86bcd71e10cbface4a479c5843c2 (::1) 0.98ms
[I 01:17:25.161 NotebookApp] Creating new notebook in
[I 01:17:26.147 NotebookApp] Kernel started: 8b702b2d-97d0-40e3-bbca-42107efd1de5
[I 01:17:27.186 NotebookApp] Adapting to protocol v5.1 for kernel 8b702b2d-97d0-40e3-bbca-42107efd1de5
And the same script ran into the Jupyter Notebook:
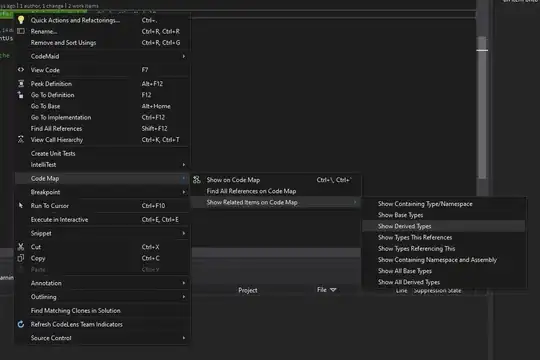
As seen, it fails and that is because it's ran by Anaconda's default Python (which doesn't have the package installed). Took a look and noticed that the jupyter-notebook executable launches (Anaconda's default) Python on jupyter-notebook-script.py (from the same dir).
Possible solutions:
1. Install the missing package(s) in the main Python
This was the 1st that came to my mind: installing PyGraphviz (and all the other required ones). Didn't try it, but it should work. The reason why I didn't try it, is because I am against polluting the main Python with packages. But, since it already contains an awful amount of site-packages, things are debatable.
2. Register the current environment Python as a kernel
I tried to make jupyter-notebook launching the current environment Python installation instead, using its configuration, or altering %CONDA_PYTHON_EXE%, but no success (note that it's my 1st time working with Jupyter). Anyway after some investigations, I realized that jupyter-notebook executable launches the Python that Jupyter is installed in. This is a common technique, and it's done by hardcoding the Python path into the executables (although strangely, looking at it with a hex editor didn't find it).
While searching, I ran into [SO]: Changing Python Executable (@Matt's answer) and from there to [ReadTheDocs.IPython]: Installing the IPython kernel, and gave that a shot:
py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> where pip
E:\Install\x64\Anaconda\Anaconda\2018.12\envs\py_064_030701_test0\Scripts\pip.exe
E:\Install\x64\Anaconda\Anaconda\2018.12\Scripts\pip.exe
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> pip freeze
certifi==2019.3.9
pygraphviz==1.5
wincertstore==0.2
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> pip install ipykernel
Collecting ipykernel
...
# Some pip useless output
...
Installing collected packages: tornado, colorama, six, ipython-genutils, decorator, traitlets, backcall, pygments, pickleshare, wcwidth, prompt-toolkit, parso, jedi, ipython, jupyter-core, python-dateutil, pyzmq, jupyter-client, ipykernel
Successfully installed backcall-0.1.0 colorama-0.4.1 decorator-4.4.0 ipykernel-5.1.0 ipython-7.4.0 ipython-genutils-0.2.0 jedi-0.13.3 jupyter-client-5.2.4 jupyter-core-4.4.0 parso-0.3.4 pickleshare-0.7.5 prompt-toolkit-2.0.9 pygments-2.3.1 python-dateutil-2.8.0 pyzmq-18.0.1 six-1.12.0 tornado-6.0.2 traitlets-4.3.2 wcwidth-0.1.7
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> where python
E:\Install\x64\Anaconda\Anaconda\2018.12\envs\py_064_030701_test0\python.exe
E:\Install\x64\Anaconda\Anaconda\2018.12\python.exe
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> python -m ipykernel install --name %CONDA_DEFAULT_ENV%
Installed kernelspec py_064_030701_test0 in C:\ProgramData\jupyter\kernels\py_064_030701_test0
After launching it, and selecting the newly created kernel (like in the image below) things went fine.
This is basically what @AndrásNagy also explained in his answer.
Although this was my 1st choice at the beginning, using current environment Python to write its metadata in a location where main Python (and other Pythons not necessarily inside Anaconda) could read it from, doesn't seem so straightforward to me (although it might be the recommended approach).
3. Install Jupyter in the current environment Python
I also thought of this from the beginning, but I didn't get to it right away because of the previous approach. I thought that Jupyter has a lot of dependencies (which it's true), but so does IPyKernel. However, now I think it's the the simplest way.
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> pip install jupyter
Collecting jupyter
...
# Some pip useless output
...
Installing collected packages: qtconsole, testpath, defusedxml, entrypoints, webencodings, bleach, mistune, MarkupSafe, jinja2, pandocfilters, attrs, pyrsistent, jsonschema, nbformat, nbconvert, Send2Trash, prometheus-client, pywinpty, terminado, notebook, widgetsnbextension, ipywidgets, jupyter-console, jupyter
Successfully installed MarkupSafe-1.1.1 Send2Trash-1.5.0 attrs-19.1.0 bleach-3.1.0 defusedxml-0.5.0 entrypoints-0.3 ipywidgets-7.4.2 jinja2-2.10 jsonschema-3.0.1 jupyter-1.0.0 jupyter-console-6.0.0 mistune-0.8.4 nbconvert-5.4.1 nbformat-4.4.0 notebook-5.7.8 pandocfilters-1.4.2 prometheus-client-0.6.0 pyrsistent-0.14.11 pywinpty-0.5.5 qtconsole-4.4.3 terminado-0.8.2 testpath-0.4.2 webencodings-0.5.1 widgetsnbextension-3.4.2
(py_064_030701_test0) [cfati@CFATI-5510-0:e:\Work\Dev\StackOverflow\q055251357]> where jupyter-notebook
E:\Install\x64\Anaconda\Anaconda\2018.12\envs\py_064_030701_test0\Scripts\jupyter-notebook.exe
E:\Install\x64\Anaconda\Anaconda\2018.12\Scripts\jupyter-notebook.exe
Needless to say that launching jupyter-notebook (notice that it's a different executable) did the trick (as installing Jupyter also registers the Python installation as a kernel).

