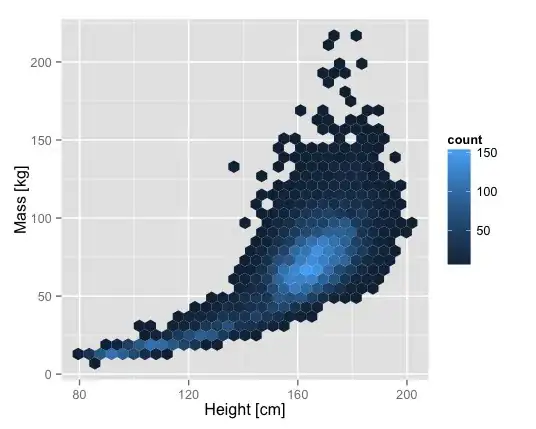
I'm generating scatter plot of the comparison of 2 variables in a data frame, where the colors of the points were determined by their type (another column in the data frame). However, there are many points that overlap each other and therefore I want an estimation of how many points there are in a given part of the grid. (for example: In case of hexbin package, one can estimate how many values there are in a given hexagon on the grid)
My question is: how can I use the hexbin package where for every group I will have another color scale, so in that way it will be possible to distinguish between the groups and get an evaluation of how many values there are.
I tried to google it but didn't find any satisfying solution. All the options that I found were focused only on the distinction between the groups.
My code so far:
ggplot(data_for_scatter_plot,aes(x=Log2FoldChange, y=TT_frequency, color=factor(Type))) +
geom_point(alpha = 0.6)
where my data_for_scatter_plot data frame is:
Gene Log2FoldChange length TT_frequency Type
ENSG00000007968 1.928153 24791 0.05623008 up_regulated
ENSG00000009724 2.209263 20711 0.05842306 down_regulated
ENSG00000010219 1.794972 53099 0.08250626 other_genes
ENSG00000053438 3.815411 2479 0.10851150 up_regulated
And the graph that I get is:
While I want to get the following graph for each group in a different color scale: