With the information I got thanks to @norok2, I got able to get a much faster solution without the loop, and to partially (i.e. only for n=2) reply the question, but not both at the same time. The solution which replies to the question is about 10 times slower:
import numpy as np
def grav_fast(p, M):
G = 6.67408*10**-2 # m³/s²T
d = p[:, :, None] - p[:, None, :]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
return (M[None, :, None]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=1)
#or return (M[None, None, :]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=2)
# (both are equivalent because d is symetric)
def grav_reply(p, M):
G = 6.67408*10**-2 # m³/s²T
d = np.tril(p[:, :, None] - p[:, None, :], -1)
d2 = np.tril((d*d).sum(axis=0), -1)
d2[d2==0] = 1
return (M[None, :, None]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=1) - \
(M[None, None, :]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=2)
# input data
system_p = np.array([[ 1., 2., 3., 4., 5., 6., 7., 9., 4., 0.],
[ 3., 5., 1., 2., 4., 5., 6., 3., 5., 8.]])
system_M = np.array([3., 5., 1., 2., 4., 5., 6., 3., 5., 8.])
# output array
l = len(system_p[0])
system_a = np.zeros(shape=(2, l))
for test in 'grav_fast', 'grav_reply':
print('\ntesting '+test)
system_a = eval(test+'(system_p, system_M)')
for i in range(l):
print('body {} mass = {}(ton), position = {}(m), '
'acceleration = [{:8.4f} {:8.4f}](m/s²)'.format(i,
system_M[i], system_p[:, i], system_a[0, i], system_a[1, i]))
grav_fast doesn't really answer the question because it makes twice the calculations, and make them also for a body attracted by itself (which causes a division by zero), but for a small system, it's still much faster than with the python's loop (break even is around 600 bodies). On the other side, grav_reply might be efficient if np.tril was designed to avoid making the calculations not needed, but it doesn't seem to be the case: A specific test with ipython showed that changing the limit diagonal in np.tril (or np.triu) didn't notably change the execution time.
In [1]: import numpy as np
In [2]: import random
In [3]: a = np.array([[random.randint(10, 99)
....: for _ in range(5)]
....: for _ in range(5)])
In [4]: %timeit np.dot(a, a)
1000000 loops, best of 3: 1.35 µs per loop
In [5]: %timeit np.tril(np.dot(a, a), 0)
100000 loops, best of 3: 17.3 µs per loop
In [6]: %timeit np.tril(np.dot(a, a), -2)
100000 loops, best of 3: 16.5 µs per loop
In [7]: a = np.array([[random.randint(10, 99)
....: for _ in range(100)]
....: for _ in range(100)])
In [8]: %timeit np.tril(a*a, 0)
10000 loops, best of 3: 56.3 µs per loop
In [9]: %timeit np.tril(a*a, -20)
10000 loops, best of 3: 61 µs per loop
In [10]: %timeit np.tril(a*a, 20)
10000 loops, best of 3: 54.7 µs per loop
In [11]: %timeit np.tril(a*a, 60)
10000 loops, best of 3: 54.5 µs per loop
Edit : Here is a performance/size graph for each algorithm
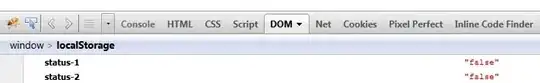
Edit : Here is the last benchmarking code I wrote:
import numpy as np
import time
import random
from matplotlib import pyplot as plt
from grav_c import grav_c, grav2_c
from numba import jit, njit
import datetime
G = 6.67408*10**-8 # m³/s²T
def grav2(p, M):
l = len(p[0])
a = np.empty(shape=(2, l))
a[:, 0] = 0
for b in range(1, l):
# computing the distance between body #b and all previous
d = p[:, b:b+1] - p[:, :b]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
# computing Newton formula : acceleration undergone by b from all previous
a[:, b] = -(M[:b] * G * d2**(-1.5) * d).sum(axis=1)
# computing Newton formula : adding for each previous, acceleration undergone by from b
a[:, :b] += M[b] * G * d2**(-1.5) * d
return a
grav2_jit = jit(grav2)
def grav(p, M):
l = len(p[0])
a = np.empty(shape=(2, l))
a[:, 0] = 0
for b in range(1, l):
# computing the distance between body #b and all previous
d = p[:, b:b+1] - p[:, :b]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
# computing Newton formula : acceleration undergone by b from all previous
a[:, b] = -(M[:b] * G / np.sqrt(d2) / d2 * d).sum(axis=1)
## a[:, b] = -(M[:b] * G * d2**(-1.5) * d).sum(axis=1)
# computing Newton formula : adding for each previous, acceleration undergone by from b
a[:, :b] += M[b] * G / np.sqrt(d2) / d2 * d
## a[:, :b] += M[b] * G * d2**(-1.5) * d
return a
grav_jit = jit(grav)
def grav2_optim1(p, M):
l = len(p[0])
a = np.empty(shape=(2, l))
a[:, 0] = 0
for b in range(1, l):
# computing the distance between body #b and all previous
d = p[:, b:b+1] - p[:, :b]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
VVV = G * d2**(-1.5)
# computing Newton formula : acceleration undergone by b from all previous
a[:, b] = -(M[:b] * VVV * d).sum(axis=1)
# computing Newton formula : adding for each previous, acceleration undergone by from b
a[:, :b] += M[b] * VVV * d
return a
grav2_optim1_jit = jit(grav2_optim1)
def grav_optim1(p, M):
l = len(p[0])
a = np.empty(shape=(2, l))
a[:, 0] = 0
for b in range(1, l):
# computing the distance between body #b and all previous
d = p[:, b:b+1] - p[:, :b]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
VVV = G / np.sqrt(d2) / d2
# computing Newton formula : acceleration undergone by b from all previous
a[:, b] = -(M[:b] * VVV * d).sum(axis=1)
# computing Newton formula : adding for each previous, acceleration undergone by from b
a[:, :b] += M[b] * VVV * d
return a
grav_optim1_jit = jit(grav_optim1)
def grav2_optim2(p, M):
l = len(p[0])
a = np.empty(shape=(2, l))
a[:, 0] = 0
for b in range(1, l):
# computing the distance between body #b and all previous
d = p[:, b:b+1] - p[:, :b]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
XXX = G * d * d2**(-1.5)
# computing Newton formula : acceleration undergone by b from all previous
a[:, b] = -(M[None, :b] * XXX).sum(axis=1)
# computing Newton formula : adding for each previous, acceleration undergone by from b
a[:, :b] += M[b] * XXX
return a
grav2_optim2_jit = jit(grav2_optim2)
def grav_optim2(p, M):
l = len(p[0])
a = np.empty(shape=(2, l))
a[:, 0] = 0
for b in range(1, l):
# computing the distance between body #b and all previous
d = p[:, b:b+1] - p[:, :b]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
XXX = G * d / np.sqrt(d2) / d2
# computing Newton formula : acceleration undergone by b from all previous
a[:, b] = -(M[None, :b] * XXX).sum(axis=1)
# computing Newton formula : adding for each previous, acceleration undergone by from b
a[:, :b] += M[b] * XXX
return a
grav_optim2_jit = jit(grav_optim2)
def grav2_vect(p, M):
d = p[:, :, None] - p[:, None, :]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
return (M[None, :, None]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=1)
grav2_vect_jit = jit(grav2_vect)
def grav_vect(p, M):
d = p[:, :, None] - p[:, None, :]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
return (M[None, :, None]*G*d/(np.sqrt(d2)*d2)[None, :, :]).sum(axis=1)
grav_vect_jit = jit(grav_vect)
# the grav*_vect_bis functions are equivalent to the grav*_vect functions because d is symetric
def grav2_vect_bis(p, M):
d = p[:, :, None] - p[:, None, :]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
return (-M[None, None, :]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=2)
grav2_vect_bis_jit = jit(grav2_vect_bis)
def grav_vect_bis(p, M):
d = p[:, :, None] - p[:, None, :]
d2 = (d*d).sum(axis=0)
d2[d2==0] = 1
return (-M[None, None, :]*G*d/(np.sqrt(d2)*d2)[None, :, :]).sum(axis=2)
grav_vect_bis_jit = jit(grav_vect_bis)
def grav2_tril(p, M):
d = np.tril(p[:, :, None] - p[:, None, :], -1)
d2 = np.tril((d*d).sum(axis=0), -1)
d2[d2==0] = 1
return (M[None, :, None]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=1) - \
(M[None, None, :]*G*d*(d2**(-1.5))[None, :, :]).sum(axis=2)
grav2_tril_jit = jit(grav2_tril)
def grav_tril(p, M):
d = np.tril(p[:, :, None] - p[:, None, :], -1)
d2 = np.tril((d*d).sum(axis=0), -1)
d2[d2==0] = 1
return (M[None, :, None]*G*d/(np.sqrt(d2)*d2)[None, :, :]).sum(axis=1) - \
(M[None, None, :]*G*d/(np.sqrt(d2)*d2)[None, :, :]).sum(axis=2)
grav_tril_jit = jit(grav_tril)
testslist = [
('grav_vect', 'c'), ('grav2_vect', 'c--'), ('grav_vect_jit', 'c:'), ('grav2_vect_jit', 'c-.'),
('grav_vect_bis', 'm'), ('grav2_vect_bis', 'm--'), ('grav_vect_bis_jit', 'm:'), ('grav2_vect_bis_jit', 'm-.'),
('grav_tril', 'y'), ('grav2_tril', 'y--'), ('grav_tril_jit', 'y:'), ('grav2_tril_jit', 'y-.'),
('grav', 'r'), ('grav2', 'r--'), ('grav_jit', 'r:'), ('grav2_jit', 'r-.'),
('grav_optim1', 'g'), ('grav2_optim1', 'g--'), ('grav_optim1_jit', 'g:'), ('grav2_optim1_jit', 'g-.'),
('grav_optim2', 'b'), ('grav2_optim2', 'b--'), ('grav_optim2_jit', 'b:'), ('grav2_optim2_jit', 'b-.'),
('grav_c', 'k'),('grav2_c', 'k--')]
class ScaleType() : pass
class LinScale(ScaleType) : pass
class LogScale(ScaleType) : pass
attempts = 8
scaletype = LogScale
scalelen = 200
scalestart = 2
scalestop = 400
# input data (Multiple datasets to repeat the tests on different data)
randlist = lambda x : [float(random.randint(10000, 99999)) for _ in range(x)]
try:
# data_file = "Here you can give an npz file name to load some presaved data.npz"
with np.load(data_file) as data:
testslist = data['testslist']
N = data['N']
timings = data['timings']
perform = data['perform']
miny = data['miny']
except NameError:
L = scalestop-scalestart
if scalelen > L:
N = np.arange(scalestart, scalestop+1, 1)
elif scaletype == LinScale:
Q = L//(scalelen-1)
R = L%(scalelen-1)
N = np.array([i for r in (range(scalestart, scalestart+Q*(scalelen-1-R), Q),
range(scalestart+Q*(scalelen-1-R), scalestop+1, Q+1)) for i in r])
elif scaletype == LogScale:
X = scalestart
G = scalestop/scalestart
I = scalelen-1
while True:
NX = I*np.log(I/np.log(G)/scalestart)/np.log(G)
if NX-X < 0.0001: break
X = NX
L0 = int(scalestart*np.power(G, X/I))
G = scalestop/(scalestart+L0)
I = scalelen-1-L0
a1 = np.array(range(I))
N = np.concatenate((range(scalestart, scalestart+L0, 1),
scalestart+L0-1+np.cumsum((0.+(scalestart+L0)*(np.exp(np.log(G)*(a1+1)/I) - np.exp(np.log(G)*a1/I))).astype(int)),
[scalestop]))
print(N)
l = len(N)
timings = np.full(l, 9999999., dtype=[(test[0], np.float64) for test in testslist])
perform = np.full(l, 9999999., dtype=[(test[0], np.float64) for test in testslist])
miny = 9999999.
accum = 0 # This is to prevent system to perform unwanted optimisations
for j in range(attempts):
for i in range(l):
L = N[i]
system_p = [np.array([randlist(L), randlist(L)]) for _ in range(100)]
system_M = [np.array( randlist(L)) for _ in range(100)]
for test in testslist:
timeref = -time.time()
system_a = eval(test[0]+'(system_p[0], system_M[0])')
accum += system_a[0, 0]
count = 1
while time.time()+timeref<0.001:
for count in range(count+1, 10*count+1):
system_a = eval(test[0]+'(system_p[count%100], system_M[count%100])')
timeref += time.time()
## print(count)
timings[test[0]][i] = min(timings[test[0]][i], timeref/count)
val = timings[test[0]][i]/(N[i]*(N[i]-1)/2)
perform[test[0]][i] = val
miny = min(val, miny)
if i%10==9: print(j, end='', flush=True)
print(flush=True)
filename = "example grav, stackoverflow "+str(datetime.datetime.now())+".npz"
print("saving data to", filename)
np.savez(filename, testslist=testslist, N=N, timings=timings, perform=perform, miny=miny)
ymin = 10**(np.floor(np.log10(miny)))
if (5*ymin<=miny): ymin *= 5
elif (2*ymin<=miny): ymin *= 2
print('ymin = {}, miny = {}\n'.format(ymin, miny))
figa, ax = plt.subplots(figsize=(24, 12))
for test in testslist:
ax.plot(N, timings[test[0]], test[1], label=test[0])
ax.set_title('numpy compared timings')
plt.xlabel('N (system size)')
plt.ylabel('timings (msec)')
plt.grid(True)
plt.legend(loc='upper left', bbox_to_anchor=(0., 1), shadow=True, ncol=7)
plt.subplots_adjust(left=0.05, bottom=0.06, right=0.98, top=0.98)
figb, bx = plt.subplots(figsize=(24, 12))
for test in testslist:
bx.plot(N, timings[test[0]], test[1], label=test[0])
bx.set_title('numpy compared timings')
plt.yscale('log')
plt.xscale('log')
plt.xlabel('N (system size)')
plt.ylabel('timings (msec)')
plt.grid(True)
plt.legend(loc='upper left', bbox_to_anchor=(0., 1), shadow=True, ncol=7)
plt.subplots_adjust(left=0.05, bottom=0.06, right=0.98, top=0.98)
figc, cx = plt.subplots(figsize=(24, 12))
for test in testslist:
cx.plot(N, perform[test[0]], test[1], label=test[0])
plt.ylim(0, 20*ymin)
cx.set_title('numpy compared performance')
plt.xlabel('N (system size)')
plt.ylabel('performance (msec)/N²')
plt.grid(True)
plt.legend(loc='upper right', bbox_to_anchor=(1., 1), shadow=True, ncol=7)
plt.subplots_adjust(left=0.05, bottom=0.06, right=0.98, top=0.98)
figd, dx = plt.subplots(figsize=(24, 12))
for test in testslist:
dx.plot(N, perform[test[0]], test[1], label=test[0])
dx.set_title('numpy compared performance')
plt.yscale('log')
plt.xscale('log')
plt.xlabel('N (system size)')
plt.ylabel('performance (msec)/N²')
plt.grid(True)
plt.legend(loc='upper right', bbox_to_anchor=(1., 1), shadow=True, ncol=7)
plt.subplots_adjust(left=0.05, bottom=0.06, right=0.98, top=0.98)
plt.show()
With it's C module
#define PY_SSIZE_T_CLEAN
#include <Python.h>
#include <numpy/arrayobject.h>
#define G 6.67408E-8L
void * failure(PyObject *type, const char *message) {
PyErr_SetString(type, message);
return NULL;
}
void * success(PyObject *var){
Py_INCREF(var);
return var;
}
static PyObject *
Py_grav_c(PyObject *self, PyObject *args)
{
PyArrayObject *p, *M;
PyObject *a;
int i, j, k;
double *pq0, *pq1, *Mq0, *Mq1, *aq0, *aq1, *p0, *p1, *a0, *a1;
if (!PyArg_ParseTuple(args, "O!O!", &PyArray_Type, &p, &PyArray_Type, &M))
return failure(PyExc_RuntimeError, "Failed to parse parameters.");
if (PyArray_DESCR(p)->type_num != NPY_DOUBLE)
return failure(PyExc_TypeError, "Type np.float64 expected for p array.");
if (PyArray_DESCR(M)->type_num != NPY_DOUBLE)
return failure(PyExc_TypeError, "Type np.float64 expected for M array.");
if (PyArray_NDIM(p)!=2)
return failure(PyExc_TypeError, "p must be a 2 dimensionnal array.");
if (PyArray_NDIM(M)!=1)
return failure(PyExc_TypeError, "M must be a 1 dimensionnal array.");
int K = PyArray_DIM(p, 0); // Number of dimensions you want
int L = PyArray_DIM(p, 1); // Number of bodies in the system
int S0 = PyArray_STRIDE(p, 0); // Normally, the arrays should be contiguous
int S1 = PyArray_STRIDE(p, 1); // But since they provide this Stride info
int SM = PyArray_STRIDE(M, 0); // I supposed they might not be (alignment)
if (PyArray_DIM(M, 0) != L)
return failure(PyExc_TypeError,
"P and M must have the same number of bodies.");
a = PyArray_NewLikeArray(p, NPY_ANYORDER, NULL, 0);
if (a == NULL)
return failure(PyExc_RuntimeError, "Failed to create output array.");
PyArray_FILLWBYTE(a, 0);
// For all bodies except first which has no previous body
for (i = 1,
pq0 = (double *)(PyArray_DATA(p)+S1),
Mq0 = (double *)(PyArray_DATA(M)+SM),
aq0 = (double *)(PyArray_DATA(a)+S1);
i < L;
i++,
*(void **)&pq0 += S1,
*(void **)&Mq0 += SM,
*(void **)&aq0 += S1
) {
// For all previous bodies
for (j = 0,
pq1 = (double *)PyArray_DATA(p),
Mq1 = (double *)PyArray_DATA(M),
aq1 = (double *)PyArray_DATA(a);
j < i;
j++,
*(void **)&pq1 += S1,
*(void **)&Mq1 += SM,
*(void **)&aq1 += S1
) {
// For all dimensions calculate deltas
long double d[K], d2 = 0, VVV, M0xVVV, M1xVVV;
for (k = 0,
p0 = pq0,
p1 = pq1;
k<K;
k++,
*(void **)&p0 += S0,
*(void **)&p1 += S0) {
d[k] = *p1 - *p0;
}
// calculate Hypotenuse squared
for (k = 0, d2 = 0; k<K; k++) {
d2 += d[k]*d[k];
}
// calculate interm. results once for each bodies pair (optimization)
VVV = G;
#define LIM 1
// if (d2<LIM) d2=LIM; // Variation on collision case
if (d2>0) VVV /= d2*sqrt(d2);
M0xVVV = *Mq0 * VVV; // anonymous intermediate result
M1xVVV = *Mq1 * VVV; // anonymous intermediate result
// For all dimensions calculate component of acceleration
for (k = 0,
a0 = aq0,
a1 = aq1;
k<K;
k++,
*(void **)&a0 += S0,
*(void **)&a1 += S0) {
*a0 += M1xVVV*d[k];
*a1 -= M0xVVV*d[k];
}
}
}
/* clean up and return the result */
return success(a);
}
static PyObject *
Py_grav2_c(PyObject *self, PyObject *args)
{
PyArrayObject *p, *M;
PyObject *a;
int i, j, k;
double *pq0, *pq1, *Mq0, *Mq1, *aq0, *aq1, *p0, *p1, *a0, *a1;
if (!PyArg_ParseTuple(args, "O!O!", &PyArray_Type, &p, &PyArray_Type, &M))
return failure(PyExc_RuntimeError, "Failed to parse parameters.");
if (PyArray_DESCR(p)->type_num != NPY_DOUBLE)
return failure(PyExc_TypeError, "Type np.float64 expected for p array.");
if (PyArray_DESCR(M)->type_num != NPY_DOUBLE)
return failure(PyExc_TypeError, "Type np.float64 expected for M array.");
if (PyArray_NDIM(p)!=2)
return failure(PyExc_TypeError, "p must be a 2 dimensionnal array.");
if (PyArray_NDIM(M)!=1)
return failure(PyExc_TypeError, "M must be a 1 dimensionnal array.");
int K = PyArray_DIM(p, 0); // Number of dimensions you want
int L = PyArray_DIM(p, 1); // Number of bodies in the system
int S0 = PyArray_STRIDE(p, 0); // Normally, the arrays should be contiguous
int S1 = PyArray_STRIDE(p, 1); // But since they provide this Stride info
int SM = PyArray_STRIDE(M, 0); // I supposed they might not be (alignment)
if (PyArray_DIM(M, 0) != L)
return failure(PyExc_TypeError,
"P and M must have the same number of bodies.");
a = PyArray_NewLikeArray(p, NPY_ANYORDER, NULL, 0);
if (a == NULL)
return failure(PyExc_RuntimeError, "Failed to create output array.");
PyArray_FILLWBYTE(a, 0);
// For all bodies except first which has no previous body
for (i = 1,
pq0 = (double *)(PyArray_DATA(p)+S1),
Mq0 = (double *)(PyArray_DATA(M)+SM),
aq0 = (double *)(PyArray_DATA(a)+S1);
i < L;
i++,
*(void **)&pq0 += S1,
*(void **)&Mq0 += SM,
*(void **)&aq0 += S1
) {
// For all previous bodies
for (j = 0,
pq1 = (double *)PyArray_DATA(p),
Mq1 = (double *)PyArray_DATA(M),
aq1 = (double *)PyArray_DATA(a);
j < i;
j++,
*(void **)&pq1 += S1,
*(void **)&Mq1 += SM,
*(void **)&aq1 += S1
) {
// For all dimensions calculate deltas
long double d[K], d2 = 0, VVV, M0xVVV, M1xVVV;
for (k = 0,
p0 = pq0,
p1 = pq1;
k<K;
k++,
*(void **)&p0 += S0,
*(void **)&p1 += S0) {
d[k] = *p1 - *p0;
}
// calculate Hypotenuse squared
for (k = 0, d2 = 0; k<K; k++) {
d2 += d[k]*d[k];
}
// calculate interm. results once for each bodies pair (optimization)
VVV = G;
#define LIM 1
// if (d2<LIM) d2=LIM; // Variation on collision case
if (d2>0) VVV *= pow(d2, -1.5);
M0xVVV = *Mq0 * VVV; // anonymous intermediate result
M1xVVV = *Mq1 * VVV; // anonymous intermediate result
// For all dimensions calculate component of acceleration
for (k = 0,
a0 = aq0,
a1 = aq1;
k<K;
k++,
*(void **)&a0 += S0,
*(void **)&a1 += S0) {
*a0 += M1xVVV*d[k];
*a1 -= M0xVVV*d[k];
}
}
}
/* clean up and return the result */
return success(a);
}
// exported functions list
static PyMethodDef grav_c_Methods[] = {
{"grav_c", Py_grav_c, METH_VARARGS, "grav_c(p, M)\n"
"\n"
"grav_c takes the positions and masses of m bodies in Newtonian attraction in a n dimensionnal universe,\n"
"and returns the accelerations each body undergoes.\n"
"input data take the for of a row of fload64 for each dimension of the position (in p) and one row for the masses.\n"
"It returns and array of the same shape as p for the accelerations."},
{"grav2_c", Py_grav2_c, METH_VARARGS, "grav_c(p, M)\n"
"\n"
"grav_c takes the positions and masses of m bodies in Newtonian attraction in a n dimensionnal universe,\n"
"and returns the accelerations each body undergoes.\n"
"input data take the for of a row of fload64 for each dimension of the position (in p) and one row for the masses.\n"
"It returns and array of the same shape as p for the accelerations."},
{NULL, NULL, 0, NULL} // pour terminer la liste.
};
static char grav_c_doc[] = "Compute attractions between n bodies.";
static struct PyModuleDef grav_c_module = {
PyModuleDef_HEAD_INIT,
"grav_c", /* name of module */
grav_c_doc, /* module documentation, may be NULL */
-1, /* size of per-interpreter state of the module,
or -1 if the module keeps state in global variables. */
grav_c_Methods
};
PyMODINIT_FUNC
PyInit_grav_c(void)
{
// I don't understand why yet, but the program segfaults without this.
import_array();
return PyModule_Create(&grav_c_module);
}