I used the ggplot2 to make a histogram. But the height of columns didn't follow the proportion of values on y-axis. Also the order of the value on y-axis isn't correct.
#This is the code I used.
data1 <- matrix(c("1", "2", "3", "4304", "456", "30"), nrow = 3, ncol = 2, dimnames = list(1:3, c("number_gRNA", "ncell")))
ggplot(data1, aes(number_gRNA, ncell)) + geom_col(width = 0.4, fill="#56B4E9", colour="black") + ggtitle("sgRNA distribution")
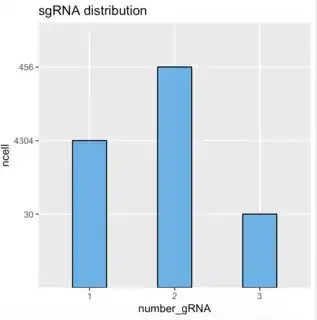
I hope the height of columns can follow the proportion with their values. Meanwhile, the values on the y-axis follow increasing order.