I got a data for comparing two genomes (column 1 and 2) and their similarity with the index indicating the number of genomes comparisons for the species:
Genome1 Genome2 % of similarity 1 in 2 % of similarity 2 in 1 Species index
1: GCF_001956585.1 GCF_002860635.1 65.2 65.0 Actinomyces_naeslundii 1
2: GCF_001937545.1 GCF_001937675.1 78.8 79.1 Actinomyces_oris 1
3: GCF_001937545.1 GCF_001937725.1 80.9 73.5 Actinomyces_oris 2
4: GCF_001937675.1 GCF_001937725.1 78.4 71.1 Actinomyces_oris 3
5: GCF_001262035.1 GCF_003130125.1 92.6 93.5 Aggregatibacter_aphrophilus 1
6: GCF_001262035.1 GCF_003252995.1 82.8 84.3 Aggregatibacter_aphrophilus 2
7: GCF_003130125.1 GCF_003252995.1 84.5 84.8 Aggregatibacter_aphrophilus 3
8: GCF_001059425.1 GCF_003130095.1 92.4 89.3 Aggregatibacter_segnis 1
9: GCF_001059425.1 GCF_003252685.1 90.1 90.1 Aggregatibacter_segnis 2
10: GCF_003130095.1 GCF_003252685.1 87.0 89.8 Aggregatibacter_segnis 3
I was trying to rotate the x-labels of a geom_points graph.
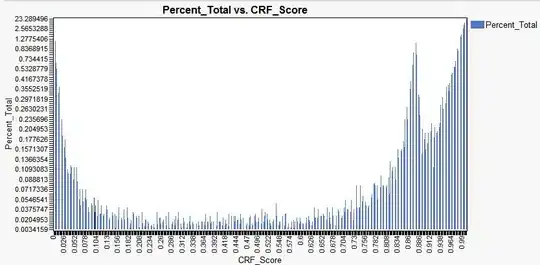
The label was successfully rotated but the graph was empty (show no data).
g <- ggplot(data = compareset, aes(x = Species, y =`% of similarity 1 in 2`, col = index))
g + geom_point()
g + theme(axis.text.x = element_text(angle = 90, hjust = 1))