What I like to do
I like to plot isochrones from multiple locations on a map so I can visually find the travel time from an arbitrary town to the closest location. It should look like a kernel density 2D plot:
library(purrr)
library(ggmap)
locations <- tibble::tribble(
~city, ~lon, ~lat,
"Hamburg", 9.992246, 53.550354,
"Berlin", 13.408163, 52.518527,
"Rostock", 12.140776, 54.088581
)
data <- map2_dfr(locations$lon, locations$lat, ~ data.frame(lon = rnorm(10000, .x, 0.8),
lat = rnorm(10000, .y, 0.7)))
ger <- c(left = min(locations$lon) - 1, bottom = min(locations$lat) - 1,
right = max(locations$lon) + 1, top = max(locations$lat) + 1)
get_stamenmap(ger, zoom = 7, maptype = "toner-lite") %>%
ggmap() +
stat_density_2d(data = data, aes(x= lon, y = lat, fill = ..level.., alpha = ..level..),
geom = "polygon") +
scale_fill_distiller(palette = "Blues", direction = 1, guide = FALSE) +
scale_alpha_continuous(range = c(0.1,0.3), guide = FALSE)
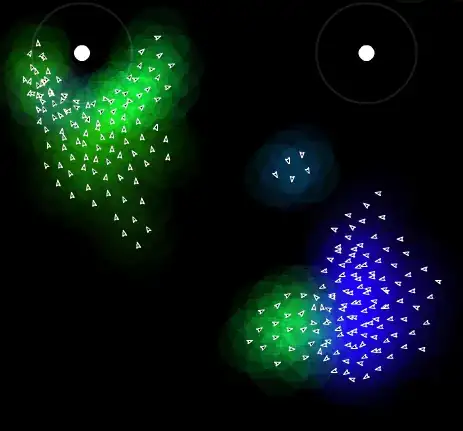
What I tried
You can easily get isochrones via osrm and plot them with leaflet. However, these isochrones are independent from each other. When I plot them they overlap each other.
library(osrm)
library(leaflet)
library(purrr)
library(ggmap)
locations <- tibble::tribble(
~city, ~lon, ~lat,
"Hamburg", 9.992246, 53.550354,
"Berlin", 13.408163, 52.518527,
"Rostock", 12.140776, 54.088581
)
isochrone <- map2(locations$lon, locations$lat,
~ osrmIsochrone(loc = c(.x, .y),
breaks = seq(0, 120, 30))) %>%
do.call(what = rbind)
isochrone@data$drive_times <- factor(paste(isochrone@data$min, "bis",
isochrone@data$max, "Minuten"))
factpal <- colorFactor("Blues", isochrone@data$drive_times, reverse = TRUE)
leaflet() %>%
setView(mean(locations$lon), mean(locations$lat), zoom = 7) %>%
addProviderTiles("Stamen.TonerLite") %>%
addPolygons(fill = TRUE, stroke = TRUE, color = "black",
fillColor = ~factpal(isochrone@data$drive_times),
weight = 0.5, fillOpacity = 0.6,
data = isochrone, popup = isochrone@data$drive_times,
group = "Drive Time") %>%
addLegend("bottomright", pal = factpal, values = isochrone@data$drive_time,
title = "Fahrtzeit")
How can I merge these isochrone so that they don't overlap?