I am completely new to programming and R, so be gentle on me ;-) - after extensive research on my problem, I still cannot figure out how to solve it...
I want to display the domains of a protein using ggplot2. I thought the best way is to use phyloseq::plot_bar() but somehow I cannot manage to do so.
I tried to compute a stacked bar plot to visualize my protein but ended up with domains in a non-specific order.
My data table looks as follows:
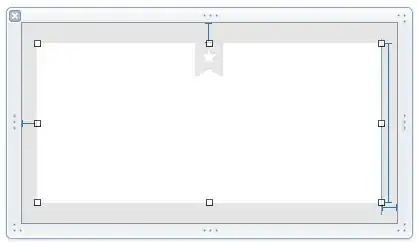
When I use ggplot to compute the bar plot, this is shown:
ggplot(my_data, aes(x="", y=length, fill=description)) +
geom_bar(stat="identity")
However, I need a linear visualization, that displays the length of each domain (description) and keeps the order according to their individual starting point set in "start".